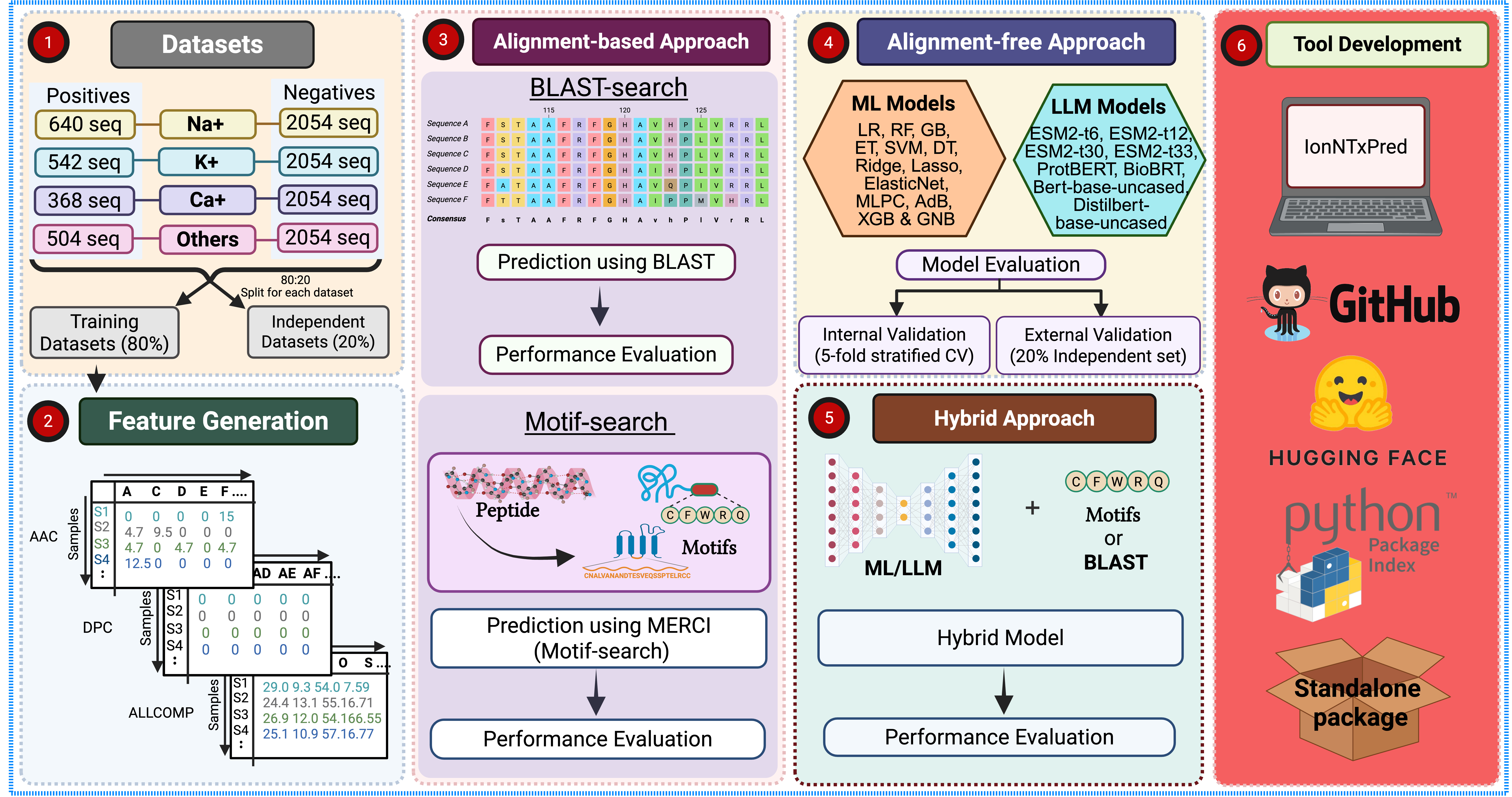
A computational framework for predicting and designing ion channel-impairing proteins using alignment-based, machine learning, and protein language model-based methods.
IonNTxPred is developed to help researchers identify proteins and peptides that modulate ion channels such as sodium (Na⁺), potassium (K⁺), calcium (Ca²⁺), and others. It integrates traditional ML models, motif discovery, and state-of-the-art protein language models (PLMs) to deliver accurate predictions and insightful biological analysis. It employs large language model for predicting toxic activity against ion channel. The final model offers Prediction, Protein-Scanning, and Design modules, implemented using protein language models.
🔗 Visit the web server for more information: IonNTxPred Web Server
🔗 Visit the Hugging Face: IonNTxPred
📖 Please cite relevant content for complete details, including the algorithm behind the approach.
Rathore et al. LLM-based Prediction and Designing of Ion Channel Impairing Proteins. #Coming Soon#
Follow these steps to replicate the core results of our paper:
# 1. Clone the repository
git clone https://github.com/raghavagps/IonNTxPred.git
cd IonNTxPred
# 2. Set up the environment (conda recommended)
conda env create -f environment.yml
conda activate IonNTxPred
# 3. Download pre-trained models
# Visit: https://webs.iiitd.edu.in/raghava/IonNTxPred/download.html
# Download the model ZIP and extract it in the root directory
# 4. See the available optiopns
python ionntxpred.py -h
# 5. Run prediction on sample input
python ionntxpred.py -i example.fasta -o output.csv -j 1 -m 1 -wd working_direcotory_path
To install IonNTxPred via PIP, run:
pip install ionntxpredTo check available options, type:
ionntxpred -hIonNTxPred is written in Python 3 and requires the following dependencies:
python=3.10.7
pytorchAdditional required packages:
pip install scikit-learn==1.5.2
pip install pandas==1.5.3
pip install numpy==1.25.2
pip install torch==2.1.0
pip install transformers==4.34.0
pip install joblib==1.4.2
pip install onnxruntime==1.15.1
Bio (Biopython): 1.81
tqdm: 4.64.1
torch: 2.6.0- Create a new Conda environment:
conda env create -f environment.yml- Activate the environment:
conda activate IonNTxPred- Due to the large size of the model file, the model directory has been compressed and uploaded.
- Download the zip file from Download Page or Hugging Face or Zenodo.
- Extract the file before using the code or model.
- place the Ectracted files inside the 'model' directory..
IonNTxPred requires BLAST+ version 2.17.0+ for running the Hybrid model.
-
Download the blast binaries file from Download Page. Please download the BLAST+ binary compatible with your operating system:
-
Linux (x64):
ncbi-blast-2.17.0+-x64-linux.tar.gz -
macOS (x64/arm64):
ncbi-blast-2.17.0+-x64-macosx.tar.gz -
Windows (x64):
ncbi-blast-2.17.0+-x64-win64.tar.gz
After downloading:
# Example for Linux
tar -xvzf ncbi-blast-2.17.0+-x64-linux.tar.gz
---
## 🔬 Classification
**IonNTxPred** classifies peptides and proteins as **ion channel impairing or non-impairing** based on their primary sequence.
🔹 **Model Options**
- **ESM2-t33**
- **Hybrid model (ESM2-t33+BLAST)**: Default Mode **
---
## 🚀 Usage
### 🔹 Minimum Usage
```bash
ionntxpred.py -hTo run an example:
ionntxpred.py -i example.fasta usage: ionntxpred.py [-h]
[-i INPUT]
[-o OUTPUT]
[-t THRESHOLD]
[-j {1,2,3,4,5}]
[-c Channel {1,2,3,4}]
[-m {1,2}]
[-d {1,2}]
[-wd WORKING DIRECTORY]| Argument | Description |
|---|---|
-i INPUT |
Input: Peptide or protein sequence (FASTA format or simple format) |
-o OUTPUT |
Output file (default: outfile.csv) |
-t THRESHOLD |
Threshold (0-1, default: 0.3) |
-j {1,2,3,4, 5} |
Job type: 1-Prediction, 2-Protein Scanning, 3-Design all possible mutants, 4- Motif Scanning, 5- BLAST Search, |
-c {1,2,3,4} |
Ion channel type: 1: Na+, 2: K+, 3: Ca+, 4: Other |
-m {1,2} |
Model selection: 1: ESM2-t33, 2: Hybrid (ESM2-t33 + BLAST) |
-wd WORKING |
Working directory for saving results |
IonNTxPred supports two formats:
- FASTA Format: (Example:
example.fasta) - Simple Format: (Example:
example.seq, each sequence on a new line)
- Results are saved in CSV format.
- If no output file is specified, results are stored in
outfile.csv.
| Job | Description |
|---|---|
| 1️⃣ Prediction | Predicts whether the input peptide/protein is an ion channel impairing or not. |
| 2️⃣ Protein Scanning | Identifies toxic regions in a protein sequence. |
| 3️⃣ Design | Generates and predicts all possible mutants. |
| 4️⃣ Motif Scanning | Identifies motifs using MERCI |
| 5️⃣ BLAST Search | Identifies toxins based BLAST hits |
| Option | Description |
|---|---|
-w {8-20} |
Window length (Protein Scan mode only, default: 12) |
-d {1,2} |
Display: 1-Ion channel impairing only, 2-All peptides (default) |
| File | Description |
|---|---|
| INSTALLATION | Installation instructions |
| LICENSE | License information |
| README.md | This file |
| IonNTxPred.py | Python program for classification |
| example.fasta | Example file (FASTA format) |
pip install ionntxpredCheck options:
ionntxpred -h🚀 Start predicting toxicity with IonNTxPred today!
🔗 Visit: IonNTxPred Web Server