-
Couldn't load subscription status.
- Fork 79
Add specimen_id attribute to the study object #2649
New issue
Have a question about this project? Sign up for a free GitHub account to open an issue and contact its maintainers and the community.
By clicking “Sign up for GitHub”, you agree to our terms of service and privacy statement. We’ll occasionally send you account related emails.
Already on GitHub? Sign in to your account
Conversation
This also adds tests and updates the DBS file, and patch (which was broken before).
And update tests, etc
Codecov Report
@@ Coverage Diff @@
## dev #2649 +/- ##
==========================================
+ Coverage 94.36% 94.38% +0.02%
==========================================
Files 166 166
Lines 19475 19563 +88
==========================================
+ Hits 18377 18465 +88
Misses 1098 1098
Continue to review full report at Codecov.
|
There was a problem hiding this comment.
Choose a reason for hiding this comment
The reason will be displayed to describe this comment to others. Learn more.
Code looks good. However, there are clear problems with performance. Also, I'm wondering if the changes affected the tags functionality via JS.
| " of these fields." % ";\n\t".join(warning_msg), | ||
| qdb.exceptions.QiitaDBWarning) | ||
|
|
||
| def unique_columns(self): |
There was a problem hiding this comment.
Choose a reason for hiding this comment
The reason will be displayed to describe this comment to others. Learn more.
This is gonna be extremely slow for large info files. Examples from the real system:
from qiita_db.metadata_template.sample_template import SampleTemplate
def unique_columns(st):
df = st.to_dataframe()
n = len(df)
return {k for k, v in df.nunique().iteritems() if v == n}
In [22]: %timeit unique_columns(SampleTemplate(550)) # moving pics
1 loop, best of 3: 662 ms per loop
In [23]: %timeit unique_columns(SampleTemplate(10317)) # AGP
1 loop, best of 3: 1min 16s per loopAs you can imagine something blocking for 1min 16s is not desirable and more in the sample info page which is pretty common and we normally expect fasts replies.
Options:
- Do everything in DB - preferred but not sure if much faster
- Do not display only unique values but all columns - and option but don't love it
- Store unique values in redis and change every time the sample id is replaced as part of the update - kind of cool to store all summaries as part of the update
- Other?
| df = self.to_dataframe() | ||
|
|
||
| n = len(df) | ||
| return {k for k, v in df.nunique().items() if v == n} |
There was a problem hiding this comment.
Choose a reason for hiding this comment
The reason will be displayed to describe this comment to others. Learn more.
items -> iteritems cause items doesn't exist in the pandas version we have in qiita
| 'message': message} | ||
| elif attribute == 'specimen_id_column': | ||
| try: | ||
| study.specimen_id_column = req_value |
There was a problem hiding this comment.
Choose a reason for hiding this comment
The reason will be displayed to describe this comment to others. Learn more.
Wondering if this step (as it has to verify that the column is unique and set the SQL values) should be a job vs. a simple set ...
There was a problem hiding this comment.
Choose a reason for hiding this comment
The reason will be displayed to describe this comment to others. Learn more.
This is only checking one column, so I don't think it will take that much time. The request should also be asynchronous from the UI so it shouldn't block any interaction. Maybe I'm overlooking something?
There was a problem hiding this comment.
Choose a reason for hiding this comment
The reason will be displayed to describe this comment to others. Learn more.
A minor fix and a question
| empty.specimen_id_column = 'foo' | ||
|
|
||
| # cleaning up the created study | ||
| qdb.study.Study.delete(empty._id) |
There was a problem hiding this comment.
Choose a reason for hiding this comment
The reason will be displayed to describe this comment to others. Learn more.
For this to always clean up, it may be worth wrapping the assertion in a try/finally
There was a problem hiding this comment.
Choose a reason for hiding this comment
The reason will be displayed to describe this comment to others. Learn more.
This can indeed become a problem. I looked into following your suggestion, however this entire test suite follows the pattern I implemented. For now I've left it as is and opened an issue with a proposed solution here #2654
| data=arguments, asjson=True) | ||
| self.assertEqual(json_decode(obs.body), {'status': 'error', 'message': | ||
| 'Study does not exist'}) | ||
| self.assertEqual(obs.code, 200) |
There was a problem hiding this comment.
Choose a reason for hiding this comment
The reason will be displayed to describe this comment to others. Learn more.
Should this be 404?
There was a problem hiding this comment.
Choose a reason for hiding this comment
The reason will be displayed to describe this comment to others. Learn more.
The handler implementing this functionality returned a 200 regardless of the status, hence I left that as is.
There was a problem hiding this comment.
Choose a reason for hiding this comment
The reason will be displayed to describe this comment to others. Learn more.
That seems surprising right? If you agree, perhaps worth an issue?
There was a problem hiding this comment.
Choose a reason for hiding this comment
The reason will be displayed to describe this comment to others. Learn more.
Yes, I agree with you. Opened here: #2655
Thanks to some further investigation by @antgonza we've concluded that we can't compute the "unique columns" in a fast way for large studies (namely the american gut project). Hence the UI now shows all the available columns and errors if a non-unique column is selected. |
|
Code looks good, a few more questions/requests:
|
Great idea, I've added a small
Yes, now it is possible.
That should be fine since this category is mainly used for mapping the tube identifiers to the Qiita sample ids. Hence if this category ceased to exist (or was deselected) the plate viewer would instead show qiita sample identifiers. For reference here's how the UI currently looks. Things to note (1) the question mark badge and the tooltip, (2) the new default in the select i.e. |
|
Thanks! Did you notice that the button beside the selector is not even, it gets cut, I guess this depends on the size of the window and the solution is to make sure the number of "bootstrap" columns is correct, can you fix? |
|
Good catch, fixed. Looks like I had incorrectly set the column width of the button. |
|
Sorry, just realized that: biocore/LabControl#175; so either we decide on a name and use it here or we remove the name from the tooltip ... |
|
I would prefer removing the name. What about "Only for use with external LIMS systems". |
|
Thanks for the feedback! |
* fix #1067 * addressing @ElDeveloper comments * fix #2512 * fix #1999 (#2514) * fix #1999 * fix template error * rm maintenance conditional * some improvements * fix #1053 * addressing @ElDeveloper comments * adding secret * WIP: EBI-ENA test (#2516) * fix Keemei (#2510) * rename preprocessed_data_id artifact-id * prints to debug * Keemei fix (#2517) * fix Keemei * delete keemei * keemei delete * rewrite_fastq=True * rmtree * add : * testing EBI * add secure * travis tests * @ElDeveloper key! * Fix typo * fix secure * rm single quote * Fix 2321 (#2522) * fix Keemei (#2510) * fix #2321 * fixing get studies * fix qiita_db * addressing @ElDeveloper comments * fixing error with skipIf * Illumina models 0318 (#2532) * fix Keemei (#2510) * updating illumina models * adding secure * Update index.rst (#2533) * fix Keemei (#2510) * Keemei fix (#2517) * fix Keemei * delete keemei * keemei delete * Update index.rst * Plugin API Doc (#2534) * fix Keemei (#2510) * plugin-api * improve format * Fix 1810 (#2523) * fix Keemei (#2510) * fix #2321 * fixing get studies * fix qiita_db * fix #1810 * adding self.ascp_pass * fix flake8 * flake8 * DOC: Cleanup of JavaScript libraries and licenses (#2536) * DOC: Cleanup of JavaScript libraries and licenses There were a few JavaScript files that were not used anywhere and we were missing a handful of license files. I've cleaned up all of that (and for consistency made all the "licence" into "license"). Fixes #2535 * DOC: Document all the JS files * BUG: Add natural sorting for data tables * adding initial download/redbiom info * adding docs * addressing @ElDeveloper comments * Fix aid str workflow (#2537) * fix Keemei (#2510) * fix int/str errors with workflows * adding a patch to fix all ints parameters * all -> any patch 64 * fix #2519 * fix #2524 * fix #2531 * fix #2529 * split qiita-cron-job into multiple commands * rm leftover docs * Update downloading.rst * fix_reference_order (#2547) * addressing @stephanieorch comments * addressing @ElDeveloper comments * Fix 2492 (#2544) * fix Keemei (#2510) * fix #2492 * flake8 * Redbiom -> redbiom @wasade * addressing @wasade comments * Update .travis.yml * Fix 2505 (#2545) * fix Keemei (#2510) * fix #2505 * fix errors * '' -> 'not provided' * Fix 2530 (#2546) * fix #2530 * fix errors * missed 1 error * more erros * fix-secure * improve available files display * fix flake8 * fix #2580 (#2584) * fix #2581 (#2583) * fixed analysis page figure (#2578) * fixed analysis page figure * removed "missing:" from null values * fix #2570 (#2586) * fix #2590 * improve sample summary speed (#2591) * fix #2562 (#2588) * fix #2562 * address @ElDeveloper comments * Update sampleTemplateVue.js * Fix 2565 (#2587) * fix #2565 * rm extra if in make-public * fix #2574 (#2585) * fix #2574 * fix error * fix ebi submission error * fix ebi assertin * adding qiimp via iframes (#2582) * adding qiimp via iframes * addressing @ElDeveloper comments * addressing #AmandaBirmingham comment * fix-test * allow to hide job via GUI (#2593) * allow to hide job via GUI * address @ElDeveloper comment * Fix upload and faq page (#2594) * fixed uplaod and faq page * fixed uplaod and faq page * fixed code and uplaod explanations * fixed code and uplaod explanations * Update upload.html * fix error * fixing bugs found during initial review * fix bug in delete column sample template (#2597) * Add qiimp xlsx (#2599) * fix-secure * improve available files display * initial code for accepting xlsx qiimp files * adding more tests and valid python 2.7 code for openpyxl * adding empty test to load_template_to_dataframe * addressing @ElDeveloper comment * rm qiimp link * Rm qiimp link (#2603) * fix-secure * improve available files display * rm qiimp link * fix #2606 * select all/none from upload * fix #2610 (#2616) * fix #2602 * fix #2601 (#2617) * fix #2601 * sphere->circle * circle_name -> var circle_name * adding ChangeLog message * TRAVIS_PULL_REQUEST -> TRAVIS_BRANCH * addressing @adswafford comment * add ignore parent for command merging scheme (#2619) * add ignore parent for command merging scheme * fix error * adding get_validator_jobs (#2620) * adding get_validator_jobs * fix error with val_job * addressing @ElDeveloper comments * fixes #2629 changed button text from Select None to Unselect All * fixes #2629 changed button text from Select None to Unselect All (#2631) * Labman minimal changes (#2633) * adding SampleInfoDBHandler * adding UserInfoDBHandlerTests * fix error * addressing @ElDeveloper comments * adding ssh backend support * rm QiitaStudySearch (#2638) * rm QiitaStudySearch * rm import search * fixing error * added paramiko * Fix 2635 (#2639) * rm QiitaStudySearch * rm import search * fixing error * fix #2635 * returning private status to preps in test_generate_study_list * addressing @ElDeveloper comments * fix #2627 (#2640) * install scp package for python * adds UsersListDBHandler (#2636) * adds UsersListDBHandler * addressing @ElDeveloper comments * rm leftover lines * testing scp directly in travis * testing scp directly in travis * testing scp directly in travis * testing scp directly in travis * testing scp directly in travis * testing scp directly in travis * fix #2632 (#2643) * fix #2632 * fixing error * fix #2608 * fix #2600 * start of edits to the upload page for SCP gui * fix #2622 (#2646) * fix #2622 * fix error * addressing @ElDeveloper comments * fix errors * Fix 2623 (#2644) * fix #2632 * fixing error * initial GUI changes * fix #2623 * sid only once * fix errors * addressing @ElDeveloper comments * fix error * @ElDeveloper comment sample_name -> sample_names * Add specimen_id attribute to the study object (#2649) * Add specimen identifier to the study table * Add specimen_id_column to study object This also adds tests and updates the DBS file, and patch (which was broken before). * ENH: Add unique_columns method to template objects * Remove maxDiff * ENH: Send response data back to the client * Make study's specimen_id_column nullable * Generalize study_tags_patch_request to be study-wide * Add Study patch handler And update tests, etc * Update UI with the new patch handlers * Add missing attribute * ENH: Add prepopulate of the UI * ENH: Add checks to prevent deletion of a specimen_id column * DOC: Delete comment * TST: Fix broken tests * PERF: Don't precompute unique columns * ENH: Add help and make specimen column deselectable * ENH: Sort column names * BUG: Fix column width for update button * DOC: Remove reference to labman * fix #2651 and partial #2652 * Fixes #2398 Strips UTF-8 characters from study titles UTF-8 characters that are not also printable ASCII characters are stripped from new study titles, as well as existing studies flagged 'public'. This is because study titles are passed as metadata to new analyses, and some analysis packages may not work well with them. * Resolving conflict with another new patch file. * Rename file * 67.py renamed to 66.py. minor edit to edit_study.html * Update study title only if UTF-8 characters were removed. * addressing @ElDeveloper comments * fix _get_valid_files * glob.glob -> glob * fix error * fix error * bringing back download_remote * improving tests * rm docstrings as requested by @ElDeveloper * fixing tests - remote_files->local_files * Scp (#2660) * fixes #2629 changed button text from Select None to Unselect All * adding ssh backend support * added paramiko * install scp package for python * testing scp directly in travis * testing scp directly in travis * testing scp directly in travis * testing scp directly in travis * testing scp directly in travis * testing scp directly in travis * addressing @ElDeveloper comments * fix _get_valid_files * glob.glob -> glob * fix error * fix error * bringing back download_remote * improving tests * rm docstrings as requested by @ElDeveloper * fixing tests - remote_files->local_files * Fixes #1802 (#2659) Password validation extended to allow only printable ASCII characters A custom validator was written to check whether a password string contains any UTF-8 characters that are not also printable ASCII characters and return false. if true. * init scp-gui * finishing scp-gui * Fixes #2398 (#2658) * Fixes #2398 Strips UTF-8 characters from study titles UTF-8 characters that are not also printable ASCII characters are stripped from new study titles, as well as existing studies flagged 'public'. This is because study titles are passed as metadata to new analyses, and some analysis packages may not work well with them. * Resolving conflict with another new patch file. * Rename file * 67.py renamed to 66.py. minor edit to edit_study.html * Update study title only if UTF-8 characters were removed. * UTF-8 stripper now processes all study-types. Code wrapped in a for loop and iterates through a list of known Study status-types; seems a little cleaner than union-ing multiple result sets. * 66.py now passes flake8 tests. * Plugin outdate version (#2664) * adding GUI changes for deprecated artifacts * fixing some tests * pull upstream dev * addressing @ElDeveloper comments * fixing errors * fixing more errors * update europeanbioinformaticsinstitute * improve ebi submission * Qsub scripts (#2667) * ADD: qsub scripts in use on qiita.ucsd.edu: * STY: flake8 * flake8 * adding changelog for this release * adding plugin updates * Dev 2553 (#2668) * minor spelling change * Migrating to per-template forbidden-words list Removed validate_invalid_column_names() from util.py, as its behavior was very specific to the needs of MetadataTemplate._clean_validate_template(). Decomposed validate_invalid_column_names() into separate methods, each validating separate concerns (invalid characters, pgsql reserved words, forbidden words). MetadataTemplate, and each Template sub-class will contain their own list of forbidden words. Currently, forbidden list is explicitly passed down call-stack; will clean-up as appropriate. * Removed explicit passing of forbidden words. Classes and tests now rely on member variable _forbidden_words to define the list of forbidden words for that Template. * Basic tests recreated for new helper methods _identify_forbidden_words_in_column_names, _identify_pgsql_reserved_words_in_column_names, and _identify_column_names_with_invalid_characters now have test cases. * Fixes #2553 Supports per-template forbidden words Support for per-template forbidden word lists. Unit tests supporting changes in util.py, base_metadata_template, etc. added. * Removed Python3 import-conditional Import import-conditional inherited from util.py * Removed docstrings for new tests. Removed docstrings for new tests. Replaced second import of quitta_db constant with a module-wide variable. * Removed PY3 from import * Modified docstring for clean_validate_template Silent dropping of 'qiita_study_id' and 'qiita_prep_id' added in docstring. * Updated comment * Modified unit-tests MetadataTemplate list of forbidden words changed to {}. Migrated forbidden_words unittest from test_base_metadata_template.py to test_sample_template.py, to give it a defined list of words to test against. Removed previously commented-out tests. * Changed _forbidden_words to be empty set {} * Removed comment * Flake8-compliant comments * SampleTemplate category names changed Category names changed from forbidden words (primer, barcode, run_prefix) to alternatives. * Added unittest to confirm silent drops Unittest confirms silent dropping of qiita_prep_id and qiita_study_id from parameter value for clean_validate_template(). * Fix Tests A few tests should be testing with assertEquals, rather than assertTrue. * Modified test to use assertEquals Test modified to use assertEquals, over assertTrue. * Replaced AssertTrue w/AssertEquals * Test modified to use assertEquals needed minor flake8 * assertItemsEqual assertion fix * fix artifact listing * checking for upload_folder * Change priority of password validation messages 'No UTF-8 characters' and 'Passwords do not match' no prioritized over password-length message. * fix scp instructions * addressing @tkosciol comment
* fix #1067 * addressing @ElDeveloper comments * fix #2512 * fix #1999 (#2514) * fix #1999 * fix template error * rm maintenance conditional * some improvements * fix #1053 * addressing @ElDeveloper comments * adding secret * WIP: EBI-ENA test (#2516) * fix Keemei (#2510) * rename preprocessed_data_id artifact-id * prints to debug * Keemei fix (#2517) * fix Keemei * delete keemei * keemei delete * rewrite_fastq=True * rmtree * add : * testing EBI * add secure * travis tests * @ElDeveloper key! * Fix typo * fix secure * rm single quote * Fix 2321 (#2522) * fix Keemei (#2510) * fix #2321 * fixing get studies * fix qiita_db * addressing @ElDeveloper comments * fixing error with skipIf * Illumina models 0318 (#2532) * fix Keemei (#2510) * updating illumina models * adding secure * Update index.rst (#2533) * fix Keemei (#2510) * Keemei fix (#2517) * fix Keemei * delete keemei * keemei delete * Update index.rst * Plugin API Doc (#2534) * fix Keemei (#2510) * plugin-api * improve format * Fix 1810 (#2523) * fix Keemei (#2510) * fix #2321 * fixing get studies * fix qiita_db * fix #1810 * adding self.ascp_pass * fix flake8 * flake8 * DOC: Cleanup of JavaScript libraries and licenses (#2536) * DOC: Cleanup of JavaScript libraries and licenses There were a few JavaScript files that were not used anywhere and we were missing a handful of license files. I've cleaned up all of that (and for consistency made all the "licence" into "license"). Fixes #2535 * DOC: Document all the JS files * BUG: Add natural sorting for data tables * adding initial download/redbiom info * adding docs * addressing @ElDeveloper comments * Fix aid str workflow (#2537) * fix Keemei (#2510) * fix int/str errors with workflows * adding a patch to fix all ints parameters * all -> any patch 64 * fix #2519 * fix #2524 * fix #2531 * fix #2529 * split qiita-cron-job into multiple commands * rm leftover docs * Update downloading.rst * fix_reference_order (#2547) * addressing @stephanieorch comments * addressing @ElDeveloper comments * Fix 2492 (#2544) * fix Keemei (#2510) * fix #2492 * flake8 * Redbiom -> redbiom @wasade * addressing @wasade comments * Update .travis.yml * Fix 2505 (#2545) * fix Keemei (#2510) * fix #2505 * fix errors * '' -> 'not provided' * Fix 2530 (#2546) * fix #2530 * fix errors * missed 1 error * more erros * fix-secure * improve available files display * fix flake8 * fix #2580 (#2584) * fix #2581 (#2583) * fixed analysis page figure (#2578) * fixed analysis page figure * removed "missing:" from null values * fix #2570 (#2586) * fix #2590 * improve sample summary speed (#2591) * fix #2562 (#2588) * fix #2562 * address @ElDeveloper comments * Update sampleTemplateVue.js * Fix 2565 (#2587) * fix #2565 * rm extra if in make-public * fix #2574 (#2585) * fix #2574 * fix error * fix ebi submission error * fix ebi assertin * adding qiimp via iframes (#2582) * adding qiimp via iframes * addressing @ElDeveloper comments * addressing #AmandaBirmingham comment * fix-test * allow to hide job via GUI (#2593) * allow to hide job via GUI * address @ElDeveloper comment * Fix upload and faq page (#2594) * fixed uplaod and faq page * fixed uplaod and faq page * fixed code and uplaod explanations * fixed code and uplaod explanations * Update upload.html * fix error * fixing bugs found during initial review * fix bug in delete column sample template (#2597) * Add qiimp xlsx (#2599) * fix-secure * improve available files display * initial code for accepting xlsx qiimp files * adding more tests and valid python 2.7 code for openpyxl * adding empty test to load_template_to_dataframe * addressing @ElDeveloper comment * rm qiimp link * Rm qiimp link (#2603) * fix-secure * improve available files display * rm qiimp link * fix #2606 * select all/none from upload * fix #2610 (#2616) * fix #2602 * fix #2601 (#2617) * fix #2601 * sphere->circle * circle_name -> var circle_name * adding ChangeLog message * TRAVIS_PULL_REQUEST -> TRAVIS_BRANCH * addressing @adswafford comment * add ignore parent for command merging scheme (#2619) * add ignore parent for command merging scheme * fix error * adding get_validator_jobs (#2620) * adding get_validator_jobs * fix error with val_job * addressing @ElDeveloper comments * fixes #2629 changed button text from Select None to Unselect All (#2631) * Labman minimal changes (#2633) * adding SampleInfoDBHandler * adding UserInfoDBHandlerTests * fix error * addressing @ElDeveloper comments * rm QiitaStudySearch (#2638) * rm QiitaStudySearch * rm import search * fixing error * Fix 2635 (#2639) * rm QiitaStudySearch * rm import search * fixing error * fix #2635 * returning private status to preps in test_generate_study_list * addressing @ElDeveloper comments * fix #2627 (#2640) * adds UsersListDBHandler (#2636) * adds UsersListDBHandler * addressing @ElDeveloper comments * rm leftover lines * fix #2632 (#2643) * fix #2632 * fixing error * fix #2608 * fix #2600 * fix #2622 (#2646) * fix #2622 * fix error * addressing @ElDeveloper comments * fix errors * Fix 2623 (#2644) * fix #2632 * fixing error * initial GUI changes * fix #2623 * sid only once * fix errors * addressing @ElDeveloper comments * fix error * @ElDeveloper comment sample_name -> sample_names * Add specimen_id attribute to the study object (#2649) * Add specimen identifier to the study table * Add specimen_id_column to study object This also adds tests and updates the DBS file, and patch (which was broken before). * ENH: Add unique_columns method to template objects * Remove maxDiff * ENH: Send response data back to the client * Make study's specimen_id_column nullable * Generalize study_tags_patch_request to be study-wide * Add Study patch handler And update tests, etc * Update UI with the new patch handlers * Add missing attribute * ENH: Add prepopulate of the UI * ENH: Add checks to prevent deletion of a specimen_id column * DOC: Delete comment * TST: Fix broken tests * PERF: Don't precompute unique columns * ENH: Add help and make specimen column deselectable * ENH: Sort column names * BUG: Fix column width for update button * DOC: Remove reference to labman * fix #2651 and partial #2652 * Fixes #2398 Strips UTF-8 characters from study titles UTF-8 characters that are not also printable ASCII characters are stripped from new study titles, as well as existing studies flagged 'public'. This is because study titles are passed as metadata to new analyses, and some analysis packages may not work well with them. * Resolving conflict with another new patch file. * Rename file * 67.py renamed to 66.py. minor edit to edit_study.html * Update study title only if UTF-8 characters were removed. * Scp (#2660) * fixes #2629 changed button text from Select None to Unselect All * adding ssh backend support * added paramiko * install scp package for python * testing scp directly in travis * testing scp directly in travis * testing scp directly in travis * testing scp directly in travis * testing scp directly in travis * testing scp directly in travis * addressing @ElDeveloper comments * fix _get_valid_files * glob.glob -> glob * fix error * fix error * bringing back download_remote * improving tests * rm docstrings as requested by @ElDeveloper * fixing tests - remote_files->local_files * UTF-8 stripper now processes all study-types. Code wrapped in a for loop and iterates through a list of known Study status-types; seems a little cleaner than union-ing multiple result sets. * 66.py now passes flake8 tests. * Additional Processing Command functionality Software.Command extended to support additional metadata regarding extra processing that may be required to merge BIOMs. * Additional Processing Command functionality Software.Command extended to support additional metadata regarding extra processing that may be required to merge BIOMs. * Fixed bug Added conditional check, where it was missing * Pass flake8 Last-minute comment did not meet flake8's approval. * Fix _method in Click _methods renamed to Qmethods * Specified Click 6.7 as a dependency * Misc cleanup Comments modified as requested. 'Q-functions' in qiita-recover-jobs have been reverted to begin with '_'. * Modified qiita-db.dbs Modified qiita-db.dbs to include additional column defined in software_command table. * Added comment metadata to new column. * Refined tests * Replaced Qtest w/_test * Reverted functions beginning w/Q to '_'.
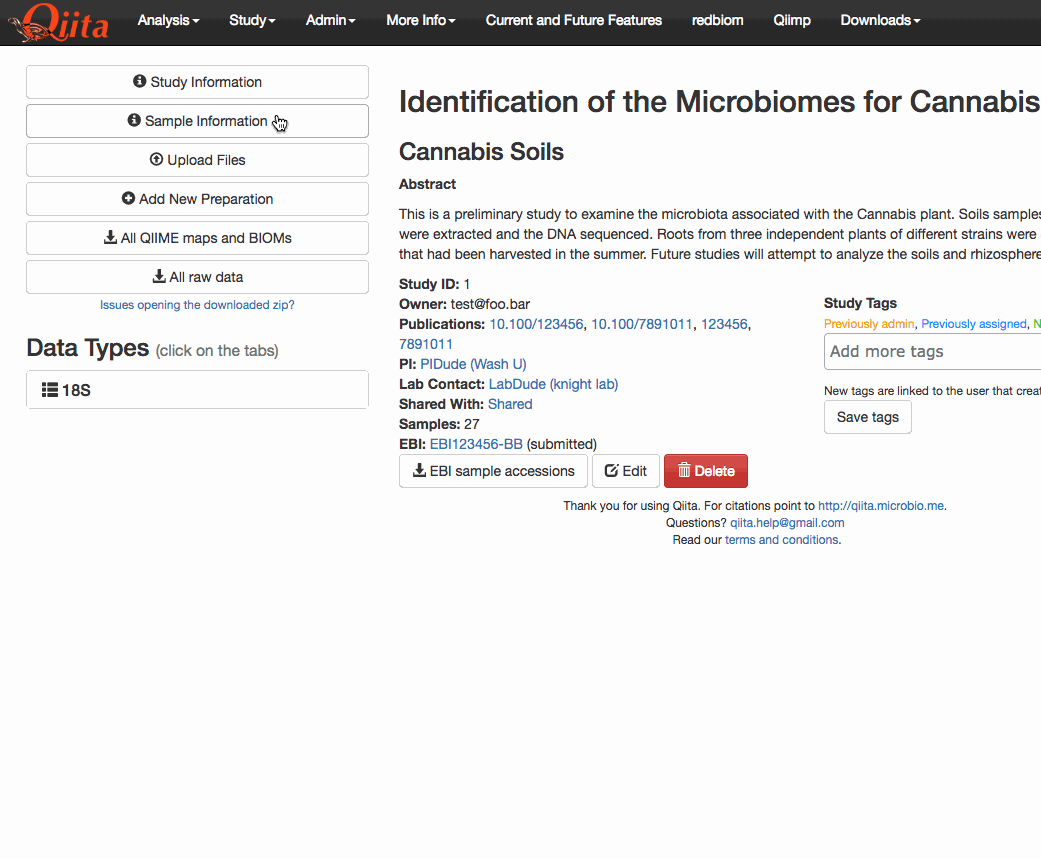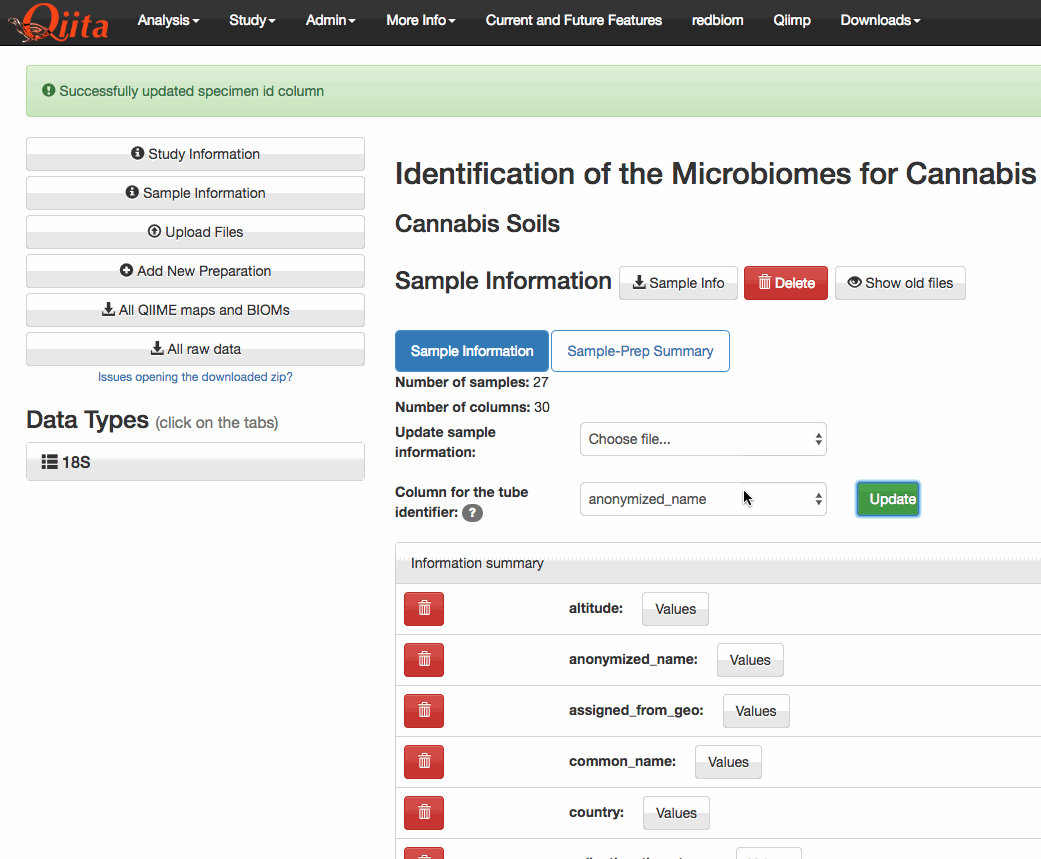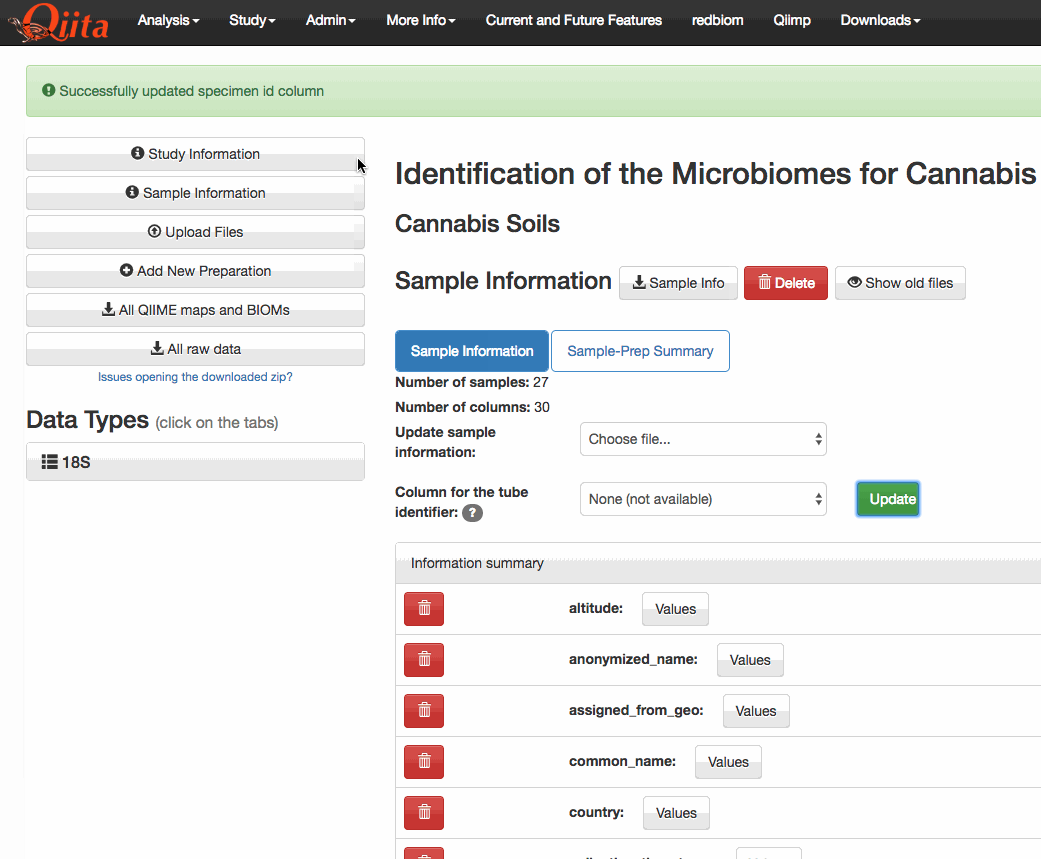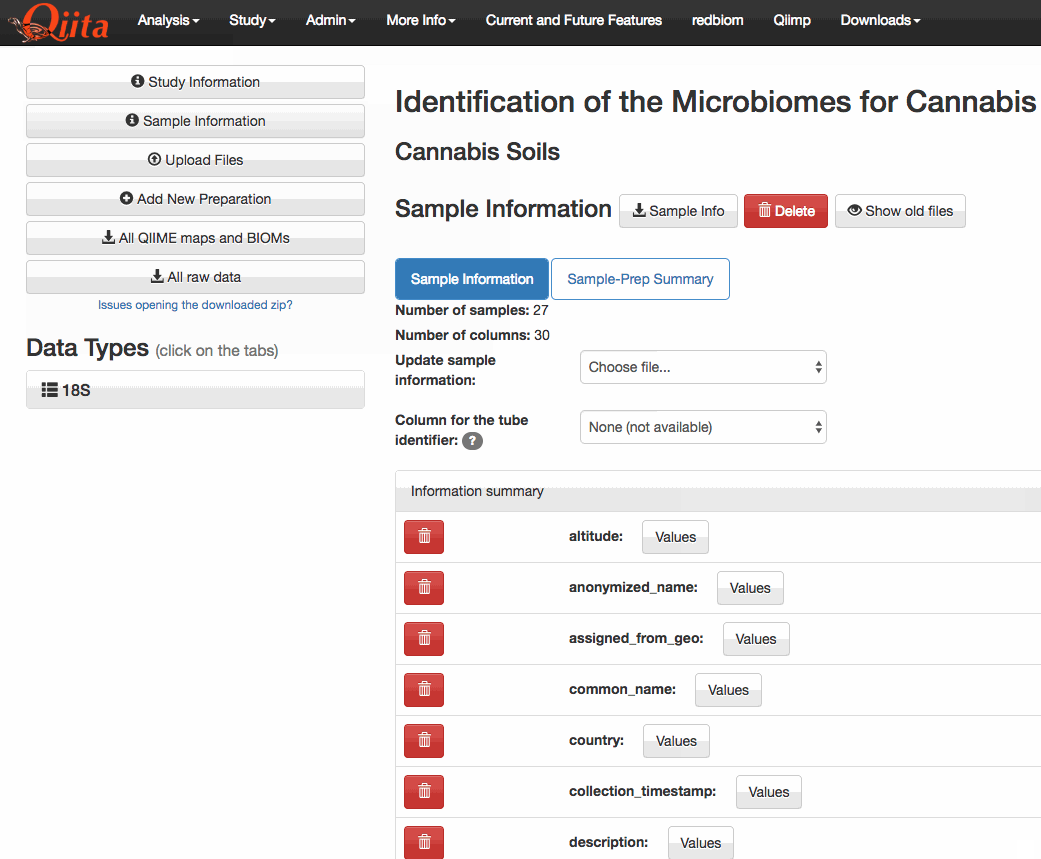
This PR adds a new attribute
specimen_id_columnto the Study object. This attribute holds the name of the column that is used to match tubes (or specimens) to sample identifiers in Qiita. The user interface has been updated to include a drop-down menu that lets the user select from any of the categories in the sample information that have unique values.I have also modified the SampleTemplate.delete_column to prevent the deletion of columns that are currently selected as a
specimen_id_column.You may notice that there are a few unrelated changes, this is because the functionality to handle PATCH requests in the Study object was limited to only operate on
tags. This PR has extended that functionality to also operate on thespecimen_id_column. While making these changes I noticed that the AJAX requests were being formatted incorrectly. The PATCH standard states that the contents be delivered via the body of the request in JSON format, not via the request's arguments. I only changed this for the requests pertaining to the study object. However, there's many other instances of this problem elsewhere in the code base. For now, I've opened #2648.The functionality itself can be seen here (note that only unique columns are selectable from the dropdown menu):
And when the user tries to delete that column:
cc @AmandaBirmingham since this is related to biocore/LabControl#273