-
Couldn't load subscription status.
- Fork 15
fixes #291 : Fix parsing of quantitation files #317
New issue
Have a question about this project? Sign up for a free GitHub account to open an issue and contact its maintainers and the community.
By clicking “Sign up for GitHub”, you agree to our terms of service and privacy statement. We’ll occasionally send you account related emails.
Already on GitHub? Sign in to your account
Conversation
… easier to read
…ml so we can catch this error? Something like this to the end of the tests: labman & LABMAN_PID=$! sleep 5 # or 10? kill $LABMAN_PID "
# Conflicts: # .travis.yml
Prevent parsing exception caused by CR newlines Interpret any values listed as "<0.00" as 0.00 Interpret any values listed as ">(some numeric value)" as "(some numeric value)" Interpret any values listed as a bunch of question marks (when you overflow the sensor; usually sample being too concentrated) as the highest quantification value in that particular file Raise an error if, after these fixes, there are any concentration values that are still NaN
There was a problem hiding this comment.
Choose a reason for hiding this comment
The reason will be displayed to describe this comment to others. Learn more.
Nothing stood out to me on a static review, this looks good!
There was a problem hiding this comment.
Choose a reason for hiding this comment
The reason will be displayed to describe this comment to others. Learn more.
One question. If answer is "yes, the user can see it via gui", let's ignore, if is not; let's open an issue to display errors via the GUI.
| # if there are any NaN concentrations left, there's a problem with the | ||
| # parsing, so throw an error | ||
| if pico_df[conc_col_name].isnull().any(): | ||
| raise ValueError("Some concentrations in pico green quantitation " |
There was a problem hiding this comment.
Choose a reason for hiding this comment
The reason will be displayed to describe this comment to others. Learn more.
Is this actually reported to user in the GUI? In other words, when you do this via the GUI and leave NaNs do you see this error?
----- As this is not part of this PR, if the answer is no; let's just open an issue.
There was a problem hiding this comment.
Choose a reason for hiding this comment
The reason will be displayed to describe this comment to others. Learn more.
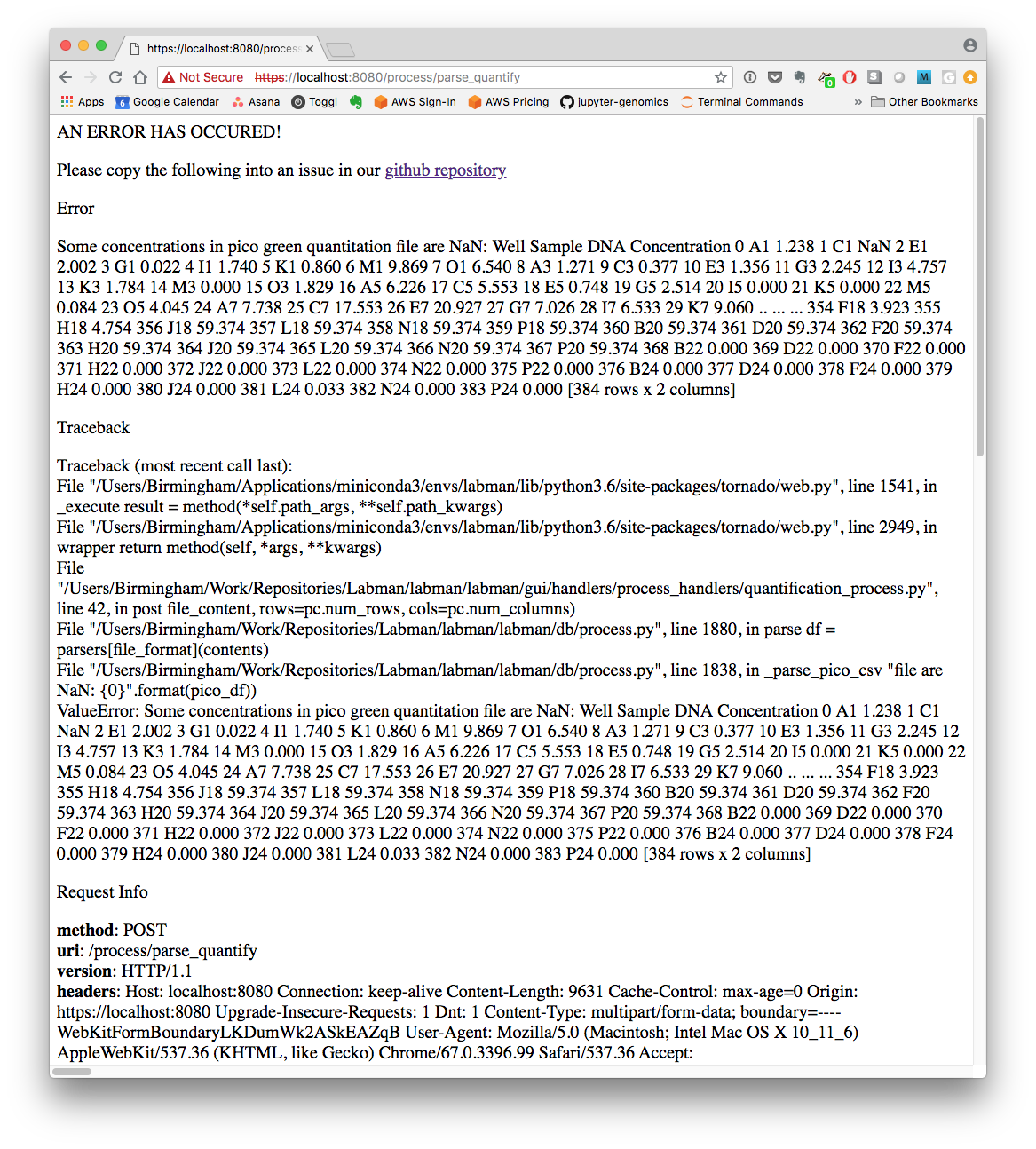
@antgonza Finally hunted this down :) The error is reported to the user in the web browser, albeit in a totally unpolished way:
Would you like me to open an issue to have it reported within the Labman page chrome, like the example below is?
There was a problem hiding this comment.
Choose a reason for hiding this comment
The reason will be displayed to describe this comment to others. Learn more.
Thanks. Let's open an issue but make it the least priority possible.
There was a problem hiding this comment.
Choose a reason for hiding this comment
The reason will be displayed to describe this comment to others. Learn more.
Will do ;)
Prevent parsing exception caused by CR newlines
Interpret any values listed as "<0.00" as 0.00
Interpret any values listed as ">some numeric value" as some numeric value
Interpret any concentrations listed as a bunch of question marks (when you overflow the sensor; usually sample being too concentrated) as the highest concentration value in that particular file
Raise an error if, after these fixes, there are any concentration values that are still NaN