-
Notifications
You must be signed in to change notification settings - Fork 2.1k
Description
2d density plots are one of the most common data-visualizations used to display flow cytometry data, and the geom_bin2d and geom_hex and geom_density_2d geoms are excellent for making these plots. However, when facetting 2d density plots, there isn't a straightforward way to set the scale such that the highest point of each plot is the same - the convention in my field.
geom_density (the 1d version) has an option to set fill = ..scaled.. which rescales all groups to a maximum of 1. Would you consider adding such an option to geom_bin2d and geom_hex (and stat_bin2d/stat_binhex)?
library(ggplot2)
library(dplyr)
library(viridis)
#2d density plot colored by level
plot.density2d_level <- diamonds %>%
ggplot(aes(x=x, y= depth)) +
stat_density_2d(aes(fill = stat(level)),
geom = "polygon",
n = 100 ,
bins = 10) +
facet_wrap(clarity~.) +
scale_fill_viridis(option = "A")
plot.density2d_level
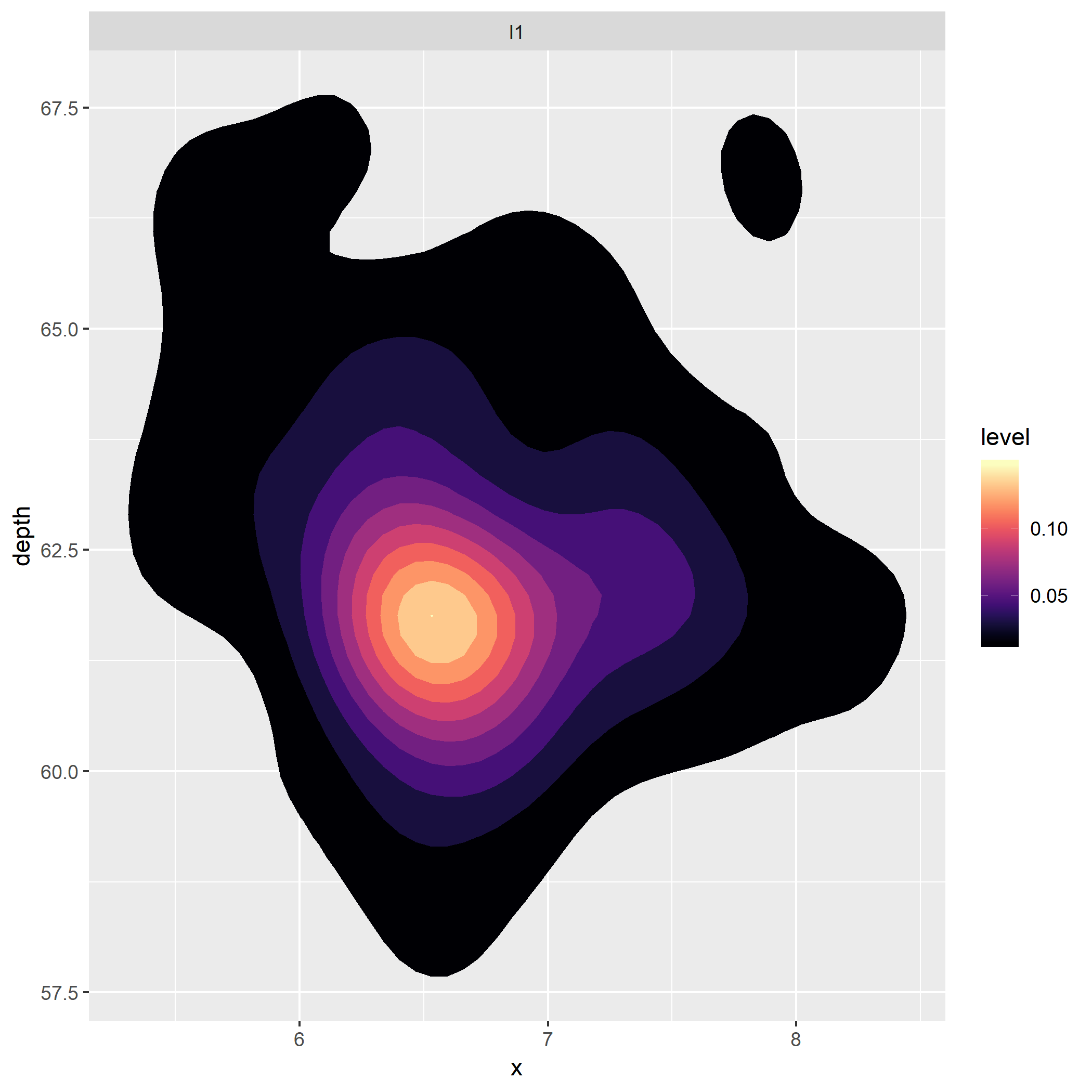
As you can see, its hard to tell where the area of highest density is in all but VVS1 and IF.
However, if I render the first panel alone, the area of highest density is very clear.
diamonds %>%
filter(clarity == "I1") %>%
ggplot(aes(x=x, y= depth)) +
stat_density_2d(aes(fill = stat(level)),
geom = "polygon",
n = 100 ,
bins = 10) +
facet_wrap(clarity~.) +
scale_fill_viridis(option = "A")
If I set the fill to stat(piece) then I'm closer to achieving the right scaling; though there are still some panels which are not using the full color palette. (This has been my temporary solution.)
plot.denisty2d_piece <- diamonds %>%
ggplot(aes(x=x, y= depth)) +
stat_density_2d(aes(fill = stat(piece)),
geom = "polygon",
n = 100,
bins = 10,
contour = T) +
facet_wrap(clarity~.) +
scale_fill_viridis(option = "A")
plot.denisty2d_piece
stat_bin2d and stat_hex also lack the ability to set the fill to a 'scaled density' such as 'ndensity' or 'ncount'
plot.bin2d <- diamonds %>%
ggplot(aes(x=x, y= depth)) +
stat_bin2d(bins = 60, aes(fill = stat(count))) +
facet_wrap(clarity~.) +
scale_fill_viridis(option = "A")
str(ggplot_build(plot.bin2d)$data[[1]])
#> 'data.frame': 2716 obs. of 20 variables:
#> $ fill : chr "#000004" "#000004" "#000004" "#000004" ...
#> $ xbin : int 27 49 38 48 54 34 42 43 48 53 ...
#> $ ybin : int 22 22 23 23 23 24 24 24 24 24 ...
#> $ value : num 1 1 1 1 1 2 1 1 1 1 ...
#> $ x : num 4.74 8.68 6.71 8.5 9.58 ...
#> $ y : num 55.5 55.5 56.1 56.1 56.1 ...
#> $ count : num 1 1 1 1 1 2 1 1 1 1 ...
#> $ density : num 0.00135 0.00135 0.00135 0.00135 0.00135 ...
#> $ PANEL : Factor w/ 8 levels "1","2","3","4",..: 1 1 1 1 1 1 1 1 1 1 ...
#> $ group : int -1 -1 -1 -1 -1 -1 -1 -1 -1 -1 ...
#> $ xmin : num 4.65 8.59 6.62 8.41 9.49 ...
#> $ xmax : num 4.83 8.77 6.8 8.59 9.67 ...
#> $ ymin : num 55.2 55.2 55.8 55.8 55.8 ...
#> $ ymax : num 55.8 55.8 56.4 56.4 56.4 ...
#> $ colour : logi NA NA NA NA NA NA ...
#> $ size : num 0.1 0.1 0.1 0.1 0.1 0.1 0.1 0.1 0.1 0.1 ...
#> $ linetype: num 1 1 1 1 1 1 1 1 1 1 ...
#> $ alpha : logi NA NA NA NA NA NA ...
#> $ width : logi NA NA NA NA NA NA ...
#> $ height : logi NA NA NA NA NA NA ...
plot.hex <- diamonds %>%
ggplot(aes(x=x, y= depth)) +
stat_binhex(bins = 60, aes(fill = stat(count))) +
facet_wrap(clarity~.) +
scale_fill_viridis(option = "A")
str(ggplot_build(plot.hex)$data[[1]])
#> 'data.frame': 3005 obs. of 10 variables:
#> $ fill : chr "#000004" "#000004" "#000004" "#000004" ...
#> $ x : num 4.74 8.5 8.68 6.62 9.49 ...
#> $ y : num 55.7 55.7 55.7 56.2 56.2 ...
#> $ density: num 0.00135 0.00135 0.00135 0.00135 0.00135 ...
#> $ count : int 1 1 1 1 1 2 1 1 1 1 ...
#> $ PANEL : Factor w/ 8 levels "1","2","3","4",..: 1 1 1 1 1 1 1 1 1 1 ...
#> $ group : int -1 -1 -1 -1 -1 -1 -1 -1 -1 -1 ...
#> $ colour : logi NA NA NA NA NA NA ...
#> $ size : num 0.5 0.5 0.5 0.5 0.5 0.5 0.5 0.5 0.5 0.5 ...
#> $ alpha : logi NA NA NA NA NA NA ...
Compare this to the 1d stat_density and stat_histogram which both provide the option to rescale each facet to a maxium of 1 using the y = stat(scaled) and y = stat(ndensity) option respectively.
plot.density <- diamonds %>%
ggplot(aes(x=x)) +
geom_density(aes(y = stat(scaled))) +
facet_wrap(clarity~.) +
scale_fill_viridis(option = "A")
plot.density
plot.histogram <- diamonds %>%
ggplot(aes(x=x)) +
geom_histogram(bins = 60, aes(y = stat(ndensity))) +
facet_wrap(clarity~.) +
scale_fill_viridis(option = "A")
plot.histogram
Would you consider adding a "scaled" statistic to stat_density2d and an "ndensity/ncount" statistic to stat_bin2d and stat_binhex?