NetST: An integrated software for large-scale haplotype network construction, visualization, and automated analytics
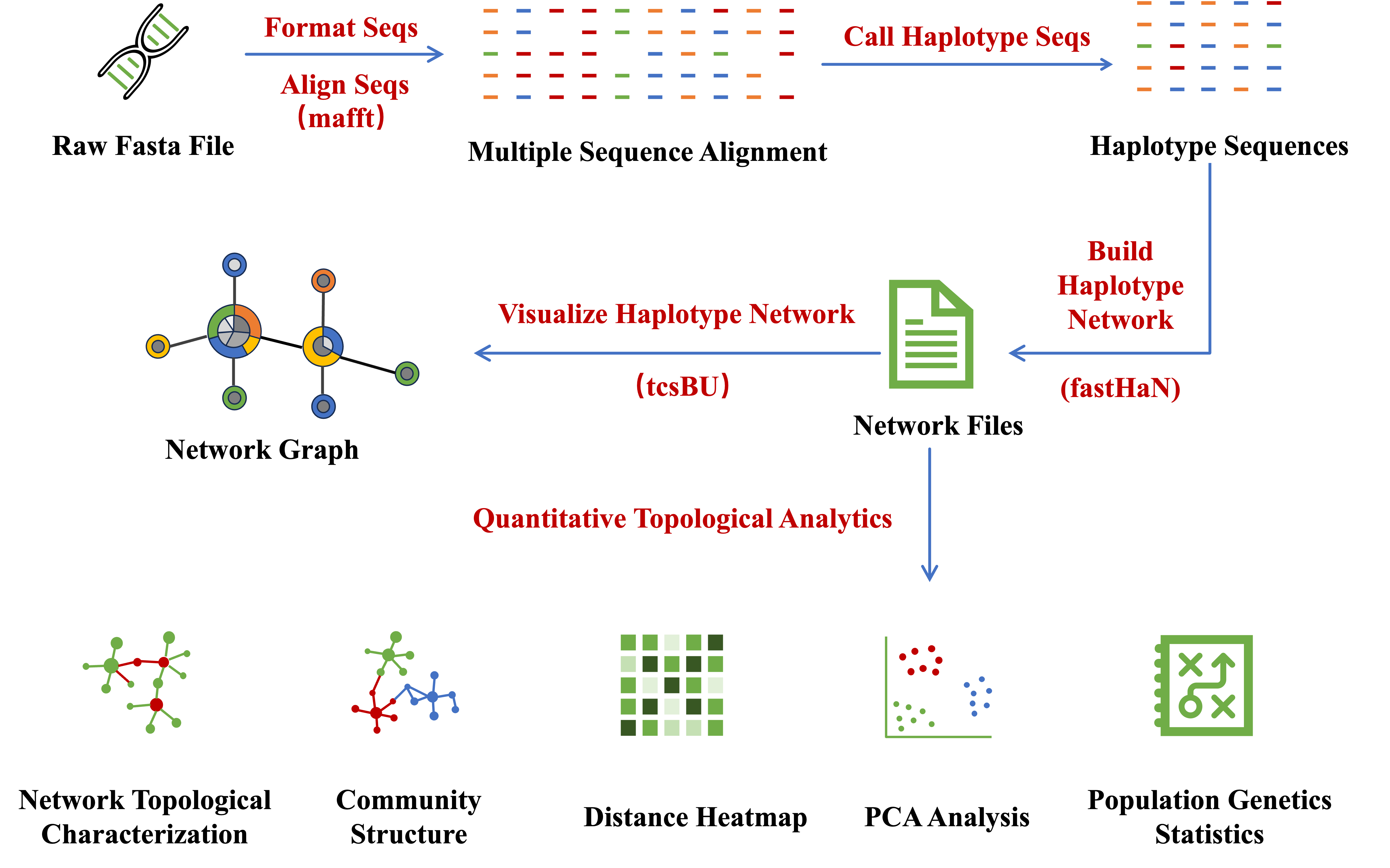
NetST is an integrated software designed to handle increasingly large and complex haplotype datasets with multiple traits in the genomics era. It enables users to efficiently calculate haplotype sequences, construct haplotype networks, visualize networks, and conduct comprehensive haplotype network analytics.
NetST implements core functionalities for haplotype network construction and visualization with a complete upstream and downstream analysis pipeline, significantly improving research efficiency. The software features innovative Quantitative Topological Analytics for network topology analysis, helping researchers better identify main groups, key hubs, and critical edges within networks.
Additionally, NetST provides dual-trait mapping and correlation analysis capabilities, allowing simultaneous visualization of associated traits within a single haplotype network and performing statistical correlation analysis on related traits, offering a more comprehensive perspective for phylogenetic relationship analysis.
Please cite the following manuscript if you use NetST:
Zhen Z, Yan Y. NetST: An Integrated Graphic Tool for Haplotype Network Construction, Visualization, and Analytics.
Additionally, please cite the dependencies when using NetST:
Chi L, Zhang X, Xue Y, Chen H. 2023. fastHaN: a fast and scalable program for constructing haplotype network for large‐sample sequences. Molecular Ecology Resources:1755-0998.13829.
Múrias Dos Santos A, Cabezas MP, Tavares AI, Xavier R, Branco M. 2016. tcsBU: a tool to extend TCS network layout and visualization. Bioinformatics 32:627–628.
Nakamura T, Yamada KD, Tomii K, Katoh K. 2018. Parallelization of MAFFT for large-scale multiple sequence alignments.Hancock J, editor. Bioinformatics 34:2490–2492.
The latest compiled version and all source code are available on GitHub. You can download the latest installation package from:
- Download the latest release package
- Extract all files to your preferred directory
- Run
netst.exeto launch the application - No additional installation required - all dependencies are included
Quick Start: For a hands-on example of how to perform haplotype network analysis using NetST, refer to the quick start tutorial: Haplotype-Network-Analysis-with-NetST
User Manual: For detailed instructions on installation, input formats, analysis modules, and output interpretation, please consult the complete manual: NetST-User-Manual
If you have any questions, suggestions, or comments about NetST, feel free to contact: yyu@scu.edu.cn,zzhen0302@163.com.