-
Notifications
You must be signed in to change notification settings - Fork 79
adding archive release #2769
New issue
Have a question about this project? Sign up for a free GitHub account to open an issue and contact its maintainers and the community.
By clicking “Sign up for GitHub”, you agree to our terms of service and privacy statement. We’ll occasionally send you account related emails.
Already on GitHub? Sign in to your account
adding archive release #2769
Conversation
Codecov Report
@@ Coverage Diff @@
## dev #2769 +/- ##
=========================================
- Coverage 94.3% 94.22% -0.09%
=========================================
Files 166 166
Lines 19980 20047 +67
=========================================
+ Hits 18843 18889 +46
- Misses 1137 1158 +21
Continue to review full report at Codecov.
|
qiita_core/tests/test_util.py
Outdated
| self.assertEqual(filepath, '') | ||
| self.assertEqual(timestamp, '') | ||
| biom_metadata_release, archive_release = get_release_info('private') | ||
| # note that we are testing not eqaul as we should have some information |
There was a problem hiding this comment.
Choose a reason for hiding this comment
The reason will be displayed to describe this comment to others. Learn more.
eqaul -> equal
There was a problem hiding this comment.
Choose a reason for hiding this comment
The reason will be displayed to describe this comment to others. Learn more.
Is there a way to hash some invariant, like metadata stored inside the BIOM file instead? Perhaps that's enough.
There was a problem hiding this comment.
Choose a reason for hiding this comment
The reason will be displayed to describe this comment to others. Learn more.
sure, added a tests to check that at least the filename is what we expect ...
There was a problem hiding this comment.
Choose a reason for hiding this comment
The reason will be displayed to describe this comment to others. Learn more.
@charles-cowart, thanks for comments; they should be addressed now.
qiita_core/tests/test_util.py
Outdated
| self.assertEqual(filepath, '') | ||
| self.assertEqual(timestamp, '') | ||
| biom_metadata_release, archive_release = get_release_info('private') | ||
| # note that we are testing not eqaul as we should have some information |
There was a problem hiding this comment.
Choose a reason for hiding this comment
The reason will be displayed to describe this comment to others. Learn more.
sure, added a tests to check that at least the filename is what we expect ...
qiita_db/archive.py
Outdated
| return dict(qdb.sql_connection.TRN.execute_fetchindex()) | ||
|
|
||
| @classmethod | ||
| def insert_features(self, merging_scheme, features): |
There was a problem hiding this comment.
Choose a reason for hiding this comment
The reason will be displayed to describe this comment to others. Learn more.
I hate to sound picky, but if insert_features is going to be a class method, can you change all instances of self to cls, just for convention's sake?
There was a problem hiding this comment.
Choose a reason for hiding this comment
The reason will be displayed to describe this comment to others. Learn more.
done.
There was a problem hiding this comment.
Choose a reason for hiding this comment
The reason will be displayed to describe this comment to others. Learn more.
thanks! Will merge as soon as testing is completed.
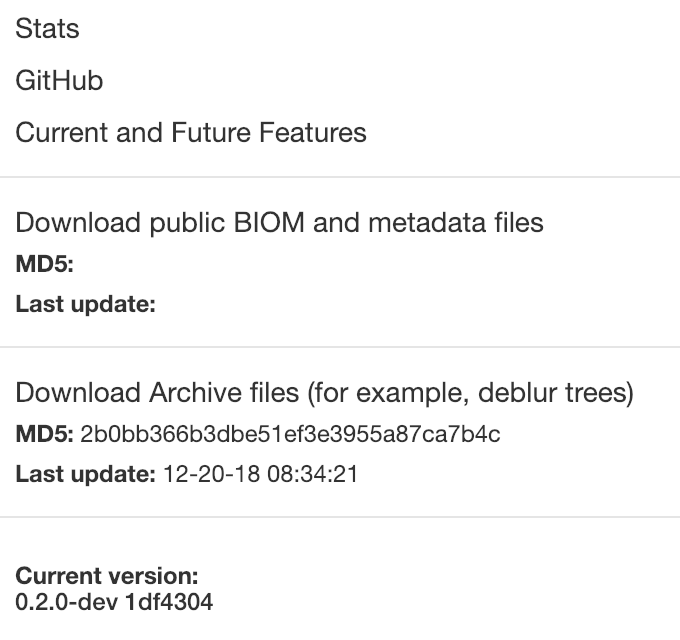
This generates the archive release; basically it allows the plugins to run the given command to generate archiving releases. Additionally we are adding a new link to download those releases.
Now, the menu looks like: