-
Notifications
You must be signed in to change notification settings - Fork 2
Description
@eprifti Hi Edi, thanks for your nice work!
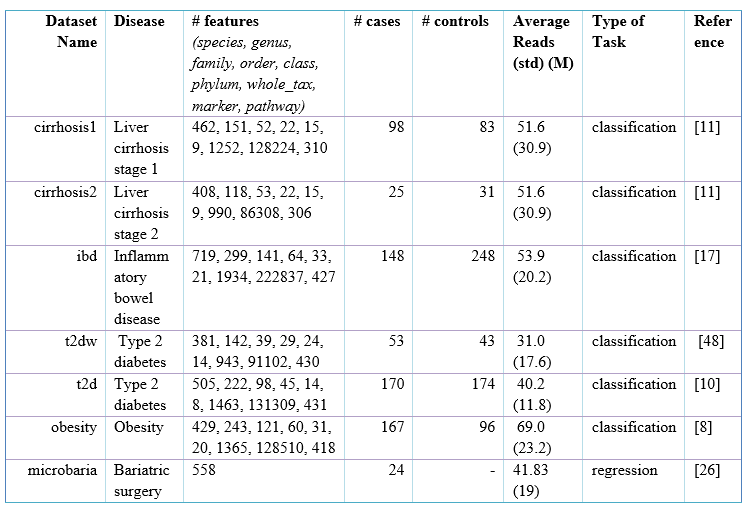
I checked the Table S1 in your paper (https://www.ncbi.nlm.nih.gov/pmc/articles/PMC7062144/#bib48), It seems that the datasets in Table S1 are not as the same datasets in this repo.
The datasets in this repo are all with 1045 features, while the datasets you've used in your paper have different number of features, and none of these datasets contain 1045 features.
It would be great if you can provideand creat those datasets you've used as benchmark datasets with pure csv format, thanks!
In addtion, I aslo fixed the installation part of predomics package, in the future, it can be very good if you can make it easy to be installed by install.package('predomics') in R. Also, I may help you convert your R predomics as Python predomics to make a Pythonic package.
install dependencies
R-version: 4.2
install.packages(c("doSNOW", "foreach", "snow", "yaml","doRNG", "gtools", "glmnet", "pROC", "viridis", "kernlab", "randomForest"))
if (!require("BiocManager", quietly = TRUE))
install.packages("BiocManager")
BiocManager::install("BioQC")
install.packages("testthat")
install.packages("roxygen2")
install.packages("reshape2")