Describe the bug
Previously, we had run into issues where the openforcefield toolkit using the OpenEye toolkit backend perceives trivalent nitrogens as stereocenters, causing issues for loading and parameterization of small molecules containing nitrogens. While we worked around this issue for nitrogens, it appears that sulfurs can also have the same problem.
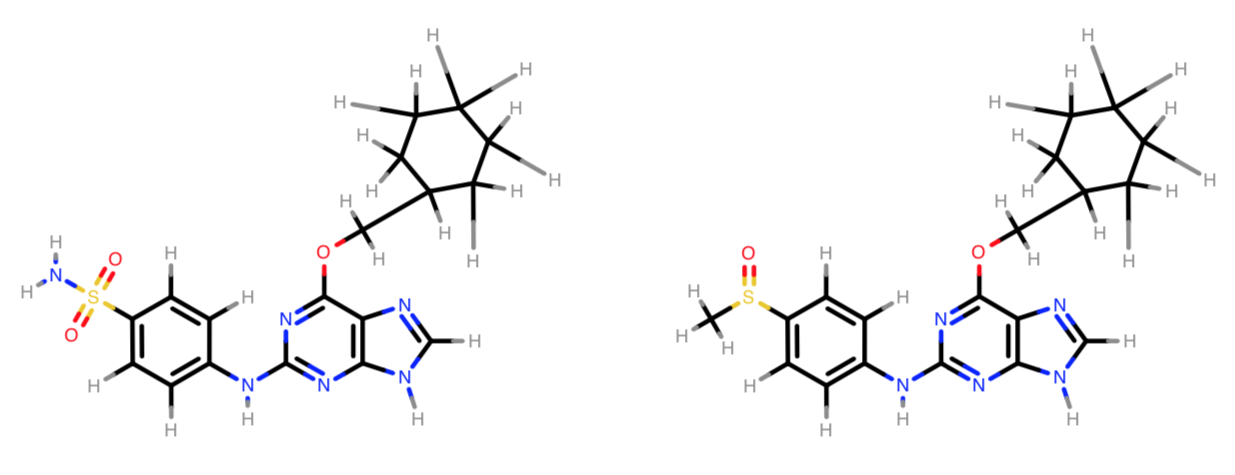
The molecule on the left has no trouble being loaded by the openforcefield toolkit, while the molecule on the right cannot be loaded without allow_undefined_stereo=True.
SDF files for both the left molecule (CDK2-no-stereo-issue.sdf) and right molecule (CDK2-stereo-issue.sdf) are provided.
To Reproduce
molecule = Molecule.from_file('../CDK2-no-stereo-issue.sdf')
molecule = Molecule.from_file('../CDK2-stereo-issue.sdf')molecules.zip
Output
With the OpenEye toolkit backend:
>>> molecule = Molecule.from_file('../CDK2-stereo-issue.sdf')
---------------------------------------------------------------------------
UndefinedStereochemistryError Traceback (most recent call last)
<ipython-input-14-354a3db15360> in <module>
----> 1 molecule = Molecule.from_file('../CDK2-stereo-issue.sdf')
~/miniconda/lib/python3.7/site-packages/openforcefield/topology/molecule.py in from_file(file_path, file_format, toolkit_registry, allow_undefined_stereo)
2989 file_path,
2990 file_format=file_format,
-> 2991 allow_undefined_stereo=allow_undefined_stereo)
2992 elif hasattr(file_path, 'read'):
2993 file_obj = file_path
~/miniconda/lib/python3.7/site-packages/openforcefield/utils/toolkits.py in from_file(self, file_path, file_format, allow_undefined_stereo)
410 from openeye import oechem
411 ifs = oechem.oemolistream(file_path)
--> 412 return self._read_oemolistream_molecules(ifs, allow_undefined_stereo, file_path=file_path)
413
414 def from_file_obj(self,
~/miniconda/lib/python3.7/site-packages/openforcefield/utils/toolkits.py in _read_oemolistream_molecules(cls, oemolistream, allow_undefined_stereo, file_path)
538 mol = cls.from_openeye(
539 oemol,
--> 540 allow_undefined_stereo=allow_undefined_stereo)
541 mols.append(mol)
542
~/miniconda/lib/python3.7/site-packages/openforcefield/utils/toolkits.py in from_openeye(oemol, allow_undefined_stereo)
757 else:
758 msg = 'Unable to make OFFMol from OEMol: ' + msg
--> 759 raise UndefinedStereochemistryError(msg)
760
761 # TODO: What other information should we preserve besides name?
UndefinedStereochemistryError: Unable to make OFFMol from OEMol: OEMol has unspecified stereochemistry. oemol.GetTitle(): 31
Problematic atoms are:
Atom atomic num: 16, name: , idx: 44, aromatic: False, chiral: True with bonds:
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 10, aromatic: True, chiral: False
bond order: 2, chiral: False to atom atomic num: 8, name: , idx: 45, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 46, aromatic: False, chiral: FalseWith the RDKit backend:
>>> molecule = Molecule.from_file('../CDK2-stereo-issue.sdf')
---------------------------------------------------------------------------
UndefinedStereochemistryError Traceback (most recent call last)
<ipython-input-28-354a3db15360> in <module>
----> 1 molecule = Molecule.from_file('../CDK2-stereo-issue.sdf')
~/miniconda/lib/python3.7/site-packages/openforcefield/topology/molecule.py in from_file(file_path, file_format, toolkit_registry, allow_undefined_stereo)
2989 file_path,
2990 file_format=file_format,
-> 2991 allow_undefined_stereo=allow_undefined_stereo)
2992 elif hasattr(file_path, 'read'):
2993 file_obj = file_path
~/miniconda/lib/python3.7/site-packages/openforcefield/utils/toolkits.py in from_file(self, file_path, file_format, allow_undefined_stereo)
1556 continue
1557 Chem.SetAromaticity(rdmol, Chem.AromaticityModel.AROMATICITY_MDL)
-> 1558 mol = self.from_rdkit(rdmol, allow_undefined_stereo=allow_undefined_stereo)
1559 mols.append(mol)
1560
~/miniconda/lib/python3.7/site-packages/openforcefield/utils/toolkits.py in from_rdkit(self, rdmol, allow_undefined_stereo)
1877 # Check for undefined stereochemistry.
1878 self._detect_undefined_stereo(rdmol, raise_warning=allow_undefined_stereo,
-> 1879 err_msg_prefix="Unable to make OFFMol from RDMol: ")
1880
1881 # Create a new openforcefield Molecule
~/miniconda/lib/python3.7/site-packages/openforcefield/utils/toolkits.py in _detect_undefined_stereo(cls, rdmol, err_msg_prefix, raise_warning)
2425 else:
2426 msg = 'Unable to make OFFMol from RDMol: ' + msg
-> 2427 raise UndefinedStereochemistryError(msg)
2428
2429 @staticmethod
UndefinedStereochemistryError: Unable to make OFFMol from RDMol: Unable to make OFFMol from RDMol: RDMol has unspecified stereochemistry. RDMol name: 31Undefined chiral centers are:
- Atom S (index 44)When specifying allow_undefined_stereo=True, the OpenEye toolkit properly gives a warning about unspecified stereochemistry:
Warning (not error because allow_undefined_stereo=True): OEMol has unspecified stereochemistry. oemol.GetTitle(): 31
Problematic atoms are:
Atom atomic num: 16, name: , idx: 44, aromatic: False, chiral: True with bonds:
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 10, aromatic: True, chiral: False
bond order: 2, chiral: False to atom atomic num: 8, name: , idx: 45, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 46, aromatic: False, chiral: False
but the RDKit does not:
>>> molecule = Molecule.from_file('../CDK2-stereo-issue.sdf', allow_undefined_stereo=True)
Warning (not error because allow_undefined_stereo=True):
The warning is not actually printed.
Computing environment (please complete the following information):
- Operating system:
osx
- Output of running
conda list
Click to expand
```
# packages in environment at /Users/choderaj/miniconda:
#
# Name Version Build Channel
alabaster 0.7.12 py_0 conda-forge
ambermini 16.16.0 7 omnia
ambertools 19.0 0 AmberMD
apbs 1.5 h1de35cc_3 schrodinger
appnope 0.1.0 py37_1000 conda-forge
arrow 0.15.4 py37_0 conda-forge
atomicwrites 1.3.0 py_0 conda-forge
attrs 19.3.0 py_0 conda-forge
autograd 1.3 py_0 conda-forge
babel 2.7.0 py_0 conda-forge
backcall 0.1.0 py_0 conda-forge
beautifulsoup4 4.8.1 py37_0 conda-forge
behave 1.2.6 pypi_0 pypi
binaryornot 0.4.4 py_1 conda-forge
bleach 3.1.0 py_0 conda-forge
blosc 1.17.0 h6de7cb9_1 conda-forge
bokeh 1.4.0 py37_0 conda-forge
boost 1.70.0 py37haf112f3_1 conda-forge
boost-cpp 1.70.0 h75728bb_2 conda-forge
bson 0.5.8 py_0 conda-forge
bzip2 1.0.8 h01d97ff_1 conda-forge
ca-certificates 2019.11.28 hecc5488_0 conda-forge
cairo 1.16.0 he1c11cd_1002 conda-forge
certifi 2019.11.28 py37_0 conda-forge
cffi 1.13.2 py37h33e799b_0 conda-forge
cftime 1.0.4.2 py37h3b54f70_0 conda-forge
chardet 3.0.4 py37_1003 conda-forge
click 7.0 py_0 conda-forge
cloudpickle 1.2.2 py_0 conda-forge
colorama 0.4.1 pypi_0 pypi
conda 4.7.12 py37_0 conda-forge
conda-build 3.18.11 py37_0 conda-forge
conda-package-handling 1.6.0 py37h01d97ff_0 conda-forge
cookiecutter 1.6.0 py37_1000 conda-forge
coverage 4.5.4 py37h0b31af3_0 conda-forge
cryptography 2.8 py37hafa8578_0 conda-forge
curl 7.65.3 h22ea746_0 conda-forge
cycler 0.10.0 py_2 conda-forge
cython 0.29.14 py37h4a8c4bd_0 conda-forge
cytoolz 0.10.1 py37h0b31af3_0 conda-forge
dask 2.7.0 py_0 conda-forge
dask-core 2.7.0 py_0 conda-forge
dbus 1.13.6 h2f22bb5_0 conda-forge
decorator 4.4.1 py_0 conda-forge
defusedxml 0.6.0 py_0 conda-forge
distributed 2.7.0 py_0 conda-forge
docutils 0.15.2 py37_0 conda-forge
entrypoints 0.3 py37_1000 conda-forge
et-xmlfile 1.0.1 pypi_0 pypi
expat 2.2.5 h4a8c4bd_1004 conda-forge
fftw3f 3.3.4 2 omnia
filelock 3.0.10 py_0 conda-forge
fontconfig 2.13.1 h6b1039f_1001 conda-forge
freemol 1.158 py37_1 schrodinger
freetype 2.10.0 h24853df_1 conda-forge
fsspec 0.5.2 py_0 conda-forge
future 0.18.2 py37_0 conda-forge
gettext 0.19.8.1 h46ab8bc_1002 conda-forge
glew 2.1.0 h4a8c4bd_0 conda-forge
glib 2.58.3 py37h577aef8_1002 conda-forge
glob2 0.7 py_0 conda-forge
h5py 2.10.0 nompi_py37h106b333_100 conda-forge
hdf4 4.2.13 h84186c3_1003 conda-forge
hdf5 1.10.5 nompi_h3e39495_1104 conda-forge
heapdict 1.0.1 py_0 conda-forge
icu 64.2 h6de7cb9_1 conda-forge
idna 2.8 py37_1000 conda-forge
imagesize 1.1.0 py_0 conda-forge
importlib_metadata 0.23 py37_0 conda-forge
ipykernel 5.1.3 py37h5ca1d4c_0 conda-forge
ipython 7.9.0 py37h5ca1d4c_0 conda-forge
ipython_genutils 0.2.0 py_1 conda-forge
ipywidgets 7.5.1 py_0 conda-forge
jdcal 1.4.1 pypi_0 pypi
jedi 0.15.1 py37_0 conda-forge
jinja2 2.10.3 py_0 conda-forge
jinja2-time 0.2.0 py_2 conda-forge
jpeg 9c h1de35cc_1001 conda-forge
jsonschema 3.1.1 py37_0 conda-forge
jupyter 1.0.0 py_2 conda-forge
jupyter_client 5.3.3 py37_1 conda-forge
jupyter_console 6.0.0 py_0 conda-forge
jupyter_core 4.5.0 py_0 conda-forge
kiwisolver 1.1.0 py37ha1b3eb9_0 conda-forge
krb5 1.16.3 hcfa6398_1001 conda-forge
libarchive 3.4.0 h1538ba8_1 conda-forge
libblas 3.8.0 14_openblas conda-forge
libcblas 3.8.0 14_openblas conda-forge
libclang 8.0.1 h770b8ee_1 conda-forge
libcurl 7.65.3 h16faf7d_0 conda-forge
libcxx 9.0.0 h89e68fa_1 conda-forge
libedit 3.1.20170329 hcfe32e1_1001 conda-forge
libffi 3.2.1 h6de7cb9_1006 conda-forge
libgcc 4.8.5 1 conda-forge
libgfortran 4.0.0 2 conda-forge
libiconv 1.15 h01d97ff_1005 conda-forge
libidn2 2.1.1 h1de35cc_0 conda-forge
liblapack 3.8.0 14_openblas conda-forge
liblief 0.9.0 h2a1bed3_1 conda-forge
libllvm8 8.0.1 h770b8ee_0 conda-forge
libnetcdf 4.7.1 nompi_he14bcb6_101 conda-forge
libopenblas 0.3.7 h4bb4525_3 conda-forge
libpng 1.6.37 h2573ce8_0 conda-forge
libsodium 1.0.17 h01d97ff_0 conda-forge
libssh2 1.8.2 hcdc9a53_2 conda-forge
libtiff 4.1.0 h3527a1b_0 conda-forge
libunistring 0.9.10 h1de35cc_0 conda-forge
libxml2 2.9.10 h53d96d6_0 conda-forge
libxslt 1.1.33 h320ff13_0 conda-forge
llvm-openmp 9.0.0 h40edb58_0 conda-forge
llvmlite 0.30.0 py37h05045ef_1 conda-forge
locket 0.2.0 py_2 conda-forge
lxml 4.4.1 py37he54a443_0 conda-forge
lz4-c 1.8.3 h6de7cb9_1001 conda-forge
lzo 2.10 h1de35cc_1000 conda-forge
markupsafe 1.1.1 py37h0b31af3_0 conda-forge
matplotlib 3.1.2 py37_0 conda-forge
matplotlib-base 3.1.2 py37h11da6c2_0 conda-forge
mdtraj 1.9.3 py37h5a8e3e4_0 conda-forge
mengine 1 h1de35cc_1 schrodinger
mistune 0.8.4 py37h0b31af3_1000 conda-forge
mmpbsa-py 16.0 pypi_0 pypi
mock 3.0.5 py37_0 conda-forge
more-itertools 7.2.0 py_0 conda-forge
mpeg_encode 1 h1de35cc_1 schrodinger
mpiplus v0.0.1 py37_1000 conda-forge
msgpack-python 0.6.2 py37ha1b3eb9_0 conda-forge
mtz2ccp4_px 1.0 hdc02c5d_3 schrodinger
nbconvert 5.6.1 py37_0 conda-forge
nbformat 4.4.0 py_1 conda-forge
ncurses 6.1 h0a44026_1002 conda-forge
netcdf4 1.5.3 py37he9269ea_0 conda-forge
networkx 2.4 py_0 conda-forge
nose 1.3.7 py37_1003 conda-forge
nose-timer 0.7.4 py_0 conda-forge
notebook 6.0.1 py37_0 conda-forge
nspr 4.20 h0a44026_1000 conda-forge
nss 3.47 hcec2283_0 conda-forge
numba 0.46.0 py37h4f17bb1_1 conda-forge
numexpr 2.7.0 py37h4f17bb1_0 conda-forge
numpy 1.17.3 py37hde6bac1_0 conda-forge
numpydoc 0.9.1 py_0 conda-forge
olefile 0.46 py_0 conda-forge
openeye-toolkits 2019.10.2 py37_0 openeye
openforcefield 0.6.0 py37_0 omnia
openforcefields 1.0.0 py37_0 omnia
openmm 7.4.0 py37_cuda101_rc_1 omnia
openmmforcefields 0.4.0+41.g6245bf3.dirty pypi_0 pypi
openmmtools 0.18.3+61.g8ca9fb0.dirty pypi_0 pypi
openmoltools 0.8.4 py37_1 omnia
openssl 1.1.1d h0b31af3_0 conda-forge
packaging 19.2 py_0 conda-forge
packmol-memgen 1.0.5rc0 pypi_0 pypi
pandas 0.25.3 py37h4f17bb1_0 conda-forge
pandoc 2.7.3 0 conda-forge
pandocfilters 1.4.2 py_1 conda-forge
parmed 3.2.0 pypi_0 pypi
parse-type 0.5.2 pypi_0 pypi
parso 0.5.1 py_0 conda-forge
partd 1.0.0 py_0 conda-forge
patsy 0.5.1 py_0 conda-forge
pcre 8.43 h4a8c4bd_0 conda-forge
pdb2pqr 2.1.1 py37_1 schrodinger
pdb4amber 1.7.dev0 pypi_0 pypi
pdbfixer 1.6 py37_0 omnia
pexpect 4.7.0 py37_0 conda-forge
pickleshare 0.7.5 py37_1000 conda-forge
pillow 6.2.1 py37hb6f49c9_0 conda-forge
pint 0.9 pypi_0 pypi
pip 19.3.1 py37_0 conda-forge
pixman 0.38.0 h01d97ff_1003 conda-forge
pkginfo 1.5.0.1 py_0 conda-forge
pluggy 0.13.0 py37_0 conda-forge
pmw 2.0.1 py37_1001 conda-forge
poyo 0.5.0 py_0 conda-forge
progressbar2 3.47.0 py_0 conda-forge
prometheus_client 0.7.1 py_0 conda-forge
prompt_toolkit 2.0.10 py_0 conda-forge
psutil 5.6.5 py37h0b31af3_0 conda-forge
ptyprocess 0.6.0 py_1001 conda-forge
py 1.8.0 py_0 conda-forge
py-lief 0.9.0 py37h6d6d4d2_1 conda-forge
pycairo 1.18.2 py37h83b1bec_0 conda-forge
pycosat 0.6.3 py37h0b31af3_1002 conda-forge
pycparser 2.19 py37_1 conda-forge
pygments 2.4.2 py_0 conda-forge
pymbar 3.0.3 py37h7eb728f_3 conda-forge
pymol 2.3.3 py37h95b93ae_0 schrodinger
pymsmt 19.0 pypi_0 pypi
pyopenssl 19.0.0 py37_0 conda-forge
pyparsing 2.4.5 py_0 conda-forge
pyqt 5.12.3 py37he22c54c_0 conda-forge
pyqt5-sip 4.19.18 pypi_0 pypi
pyqtwebengine 5.12.1 pypi_0 pypi
pyrsistent 0.15.5 py37h0b31af3_0 conda-forge
pysocks 1.7.1 py37_0 conda-forge
pytables 3.6.1 py37h6f8395a_0 conda-forge
pytest 5.2.2 py37_0 conda-forge
pytest-cov 2.8.1 py_0 conda-forge
pytest-runner 5.2 py_0 conda-forge
python 3.7.3 h93065d6_1 conda-forge
python-dateutil 2.8.1 py_0 conda-forge
python-libarchive-c 2.9 py37_0 conda-forge
python-utils 2.3.0 py_1 conda-forge
python.app 1.2 py37h0b31af3_1201 conda-forge
pytraj 2.0.5 pypi_0 pypi
pytz 2019.3 py_0 conda-forge
pyyaml 5.1.2 py37h0b31af3_0 conda-forge
pyzmq 18.1.0 py37h4bf09a9_0 conda-forge
qt 5.12.5 h1b46049_0 conda-forge
qtconsole 4.5.5 py_0 conda-forge
rdkit 2019.09.1 py37h3471270_0 conda-forge
readline 8.0 hcfe32e1_0 conda-forge
rednose 1.3.0 pypi_0 pypi
requests 2.22.0 py37_1 conda-forge
rigimol 1.3 2 schrodinger
ripgrep 11.0.2 h01d97ff_3 conda-forge
ruamel_yaml 0.15.71 py37h1de35cc_1000 conda-forge
sander 16.0 pypi_0 pypi
scipy 1.3.2 py37h82752d6_0 conda-forge
seaborn 0.9.0 py_2 conda-forge
send2trash 1.5.0 py_0 conda-forge
setuptools 41.6.0 py37_1 conda-forge
six 1.13.0 py37_0 conda-forge
smirnoff99frosst 1.1.0 py37_1 omnia
snowballstemmer 2.0.0 py_0 conda-forge
sortedcontainers 2.1.0 py_0 conda-forge
soupsieve 1.9.4 py37_0 conda-forge
sphinx 2.2.1 py_0 conda-forge
sphinxcontrib-applehelp 1.0.1 py_0 conda-forge
sphinxcontrib-devhelp 1.0.1 py_0 conda-forge
sphinxcontrib-htmlhelp 1.0.2 py_0 conda-forge
sphinxcontrib-jsmath 1.0.1 py_0 conda-forge
sphinxcontrib-qthelp 1.0.2 py_0 conda-forge
sphinxcontrib-serializinghtml 1.1.3 py_0 conda-forge
sqlite 3.30.1 h93121df_0 conda-forge
statsmodels 0.10.1 py37h3b54f70_1 conda-forge
tblib 1.4.0 py_0 conda-forge
termcolor 1.1.0 py_2 conda-forge
terminado 0.8.2 py37_0 conda-forge
testpath 0.4.4 py_0 conda-forge
tinydb 3.15.1 py_0 conda-forge
tk 8.6.9 x11tk0_2000 schrodinger
toml 0.10.0 py_0 conda-forge
toolz 0.10.0 py_0 conda-forge
tornado 6.0.3 py37h0b31af3_0 conda-forge
tqdm 4.37.0 py_0 conda-forge
traitlets 4.3.3 py37_0 conda-forge
urllib3 1.25.7 py37_0 conda-forge
wcwidth 0.1.7 py_1 conda-forge
webencodings 0.5.1 py_1 conda-forge
wget 1.20.1 h33e2efd_0 conda-forge
wheel 0.33.6 py37_0 conda-forge
whichcraft 0.6.1 py_0 conda-forge
widgetsnbextension 3.5.1 py37_0 conda-forge
xmltodict 0.12.0 py_0 conda-forge
xz 5.2.4 h1de35cc_1001 conda-forge
yaml 0.1.7 h1de35cc_1001 conda-forge
zeromq 4.3.2 h6de7cb9_2 conda-forge
zict 1.0.0 py_0 conda-forge
zipp 0.6.0 py_0 conda-forge
zlib 1.2.11 h0b31af3_1006 conda-forge
zstd 1.4.3 he7fca8b_0 conda-forge
```
Additional context
cc: #382 #146
