#Apache Spark OpenCPU Executor (ROSE)
ROSE is an Apache Spark package offering access to the full scientific computing power of the R programming language to Spark batch and streaming applications on the JVM. This library is built on top of the opencpu-r-executor library, a lightweight solution for integrating R analytics executed on the OpenCPU server into any application running on the JVM.
Where Apache SparkR lets data scientists use Spark from R, ROSE is designed to let Scala and Java developers use R from Spark.
The popular Apache SparkR package provides a lightweight front-end for data scientists to use Apache Spark from R. This approach is ideally suited to investigative analytics, such as ad-hoc and exploratory analysis at scale.
The ROSE library attempts to provide the same R analytics capabilities
available to Apache SparkR applications within traditional Spark applications
on the JVM. It does this by exposing new analyze operations that execute R
analytics on compatible RDDs. This new facility is designed primarily for
operational analytics and can be used alongside Spark core, SQL, Streaming,
MLib and GraphX.
If you need to query R machine-learning models, score R prediction models or leverage any other aspect of the R library within your Spark applications on the JVM then the ROSE library may be for you.
A number of example applications are provided to demonstrate the use of the ROSE library to deliver R analytics capabilities within any Spark solution.
- Hello, World! [ Scala ][ Java ]
- Batch Predictive Scoring Engine [ Scala ][ Java ]
- Streaming Predictive Scoring Engine [ Scala ][ Java ]
- Twitter Sentiment Analysis [ Scala ][ Java ]
libraryDependencies += "io.onetapbeyond" %% "opencpu-spark-executor_2.10" % "1.0"
compile 'io.onetapbeyond:opencpu-spark-executor_2.10:1.0'
Include the ROSE package in your Spark application using spark-shell, or spark-submit. For example:
$SPARK_HOME/bin/spark-shell --packages io.onetapbeyond:opencpu-spark-executor_2.10:1.0
This library exposes new analyze transformations on Spark RDDs of type
RDD[OCPUTask]. The following sections demonstrate how to use these new
RDD operations to execute R analytics directly within Spark batch and
streaming applications on the JVM.
See the documentation
on the underlying opencpu-r-executor library for details on building
OCPUTask and handling OCPUResult.
For this example we assume an input dataRDD, then transform it to generate
an RDD of type RDD[OCPUTask]. In this example each OCPUTask represents a
fraud score prediction to be generated by the R function fraud::score when
the RDD is eventually evaluated.
import io.onetapbeyond.opencpu.spark.executor.R._
import io.onetapbeyond.opencpu.r.executor._
val rTaskRDD = dataRDD.map(data => {
OCPU.R()
.pkg("fraud")
.function("score")
.input(data.asInput())
.library()
})
The set of OCPUTask within rTaskRDD can be scheduled for
processing by calling the new analyze operation provided by ROSE
on the RDD:
val rResultRDD = rTaskRDD.analyze
When rTaskRDD.analyze is evaluated by Spark the resultant rResultRDD
is of type RDD[OCPUResult]. The fraud prediction score for the original
OCPUTask are available within these OCPUResult. These values can be
optionally cached, further processed or persisted per the needs of your
Spark application.
Note, the use here of the R function fraud::score is simply representative
of any R function or script available within the full set of R packages
available on CRAN R,
Bioconductor or on github.
For this example we assume an input stream dataStream, then transform
it to generate a new stream with underlying RDDs of type RDD[OCPUTask].
In this example each OCPUTask represents a fraud score prediction to
be generated by the R function fraud::score when the stream is
eventually evaluated.
import io.onetapbeyond.opencpu.spark.executor.R._
import io.onetapbeyond.opencpu.r.executor._
val rTaskStream = dataStream.transform(rdd => {
rdd.map(data => {
OCPU.R()
.pkg("fraud")
.function("score")
.input(data.asInput())
.library()
})
})
The set of OCPUTask within rTaskStream can be scheduled for processing
by calling the new analyze operation provided by ROSE on each RDD within
the stream:
val rResultStream = rTaskStream.transform(rdd => rdd.analyze)
When rTaskStream.transform is evaluated by Spark the resultant
rResultStream has underlying RDDs of type RDD[OCPUResult]. The fraud
prediction score for the original OCPUTask are available within these
OCPUResult. These values can be optionally cached, further processed
or persisted per the needs of your Spark application.
Note, the use here of the R function fraud::score is simply
representative of any R function or script available within the full
set of R packages available on CRAN R,
Bioconductor or on github.
To understand how ROSE delivers the full scientific computing power of the R programming language to Spark applications on the JVM the following sections compare and constrast the deployment of traditional Scala, Java, Python and SparkR applications with Spark applications powered by the ROSE library.
The principal deployment requirement when working with ROSE is that your
Spark cluster have access to one or more
OpenCPU servers. Deployment
options for those servers in the context of Spark are discussed in
Application Deployment sections 3. and 4. that follow below.
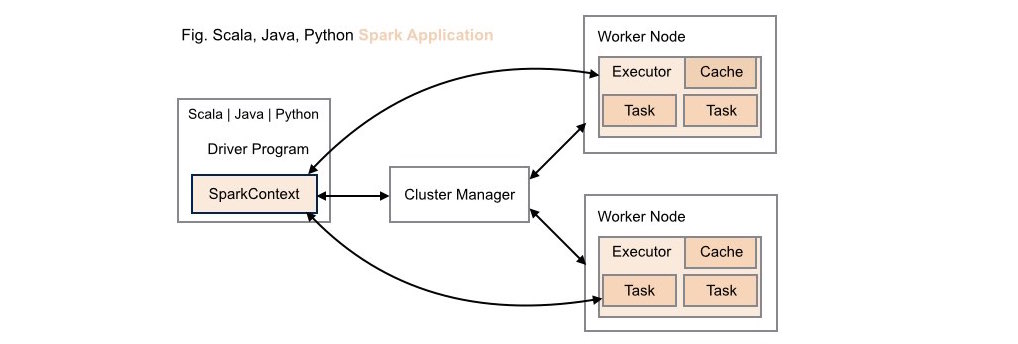
####1. Traditional Scala | Java | Python Spark Application Deployment
Without ROSE library support, neither data scientists nor application developers have access to R's analytic capabilities within these types of application deployments.
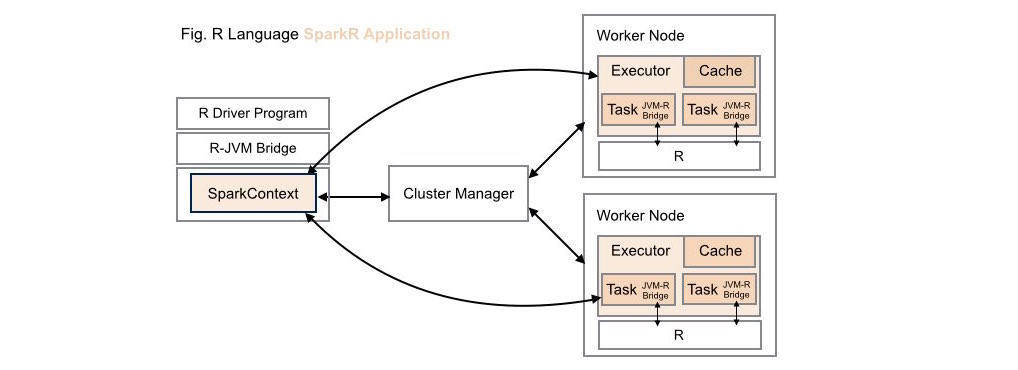
####2. Traditional SparkR Application Deployment
While data scientists can leverage the computing power of Spark within R applications in these types of application deployments, these same R capabilities are not available to Scala, Java or Python developers.
Note, when working with Apache SparkR, the R runtime environment must be installed locally on each worker node on your cluster.
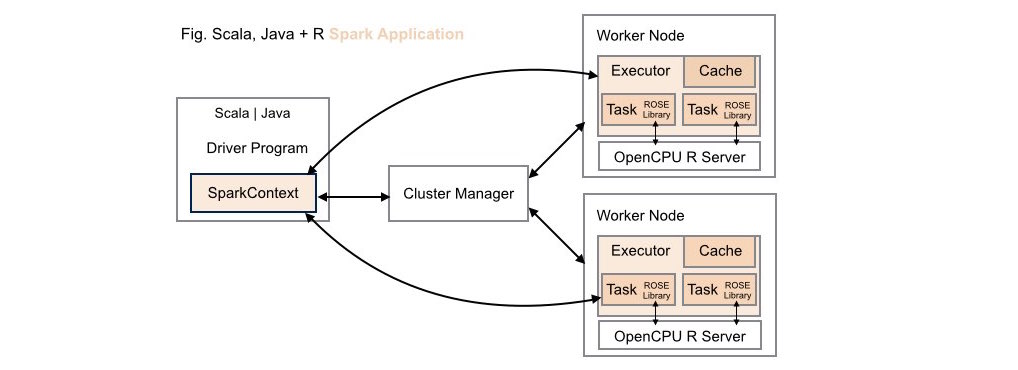
####3. Scala | Java + R (ROSE) Spark Application Deployment
Both data scientists and application developers working in either Scala or Java can leverage the full power of R using the ROSE library within these types of application deployments.
Using this deployment configuration, each worker node on the Spark cluster
has its own dedicated OpenCPU server installed locally on the node. This
configuration delivers optimal runtime throughput on the cluster. Note, this
deployment configuration mirrors the configuration required by Apache
SparkR, where the R runtime environment must be installed locally on each
worker node on the cluster.
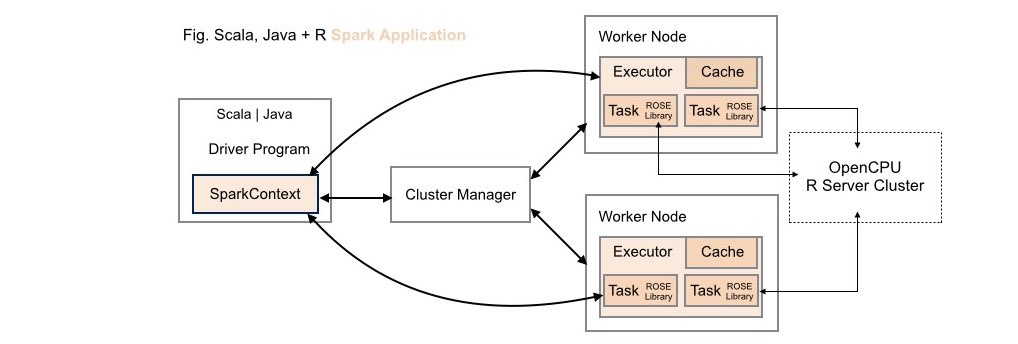
####4. Scala | Java + R (ROSE) Spark Application Deployment (Alternative)
Both data scientists and application developers working in either Scala or Java can leverage the full power of R using the ROSE library within these types of application deployments.
Using this deployment configuration, a cluster of one or more
OpenCPU servers is maintained external to the Spark cluster. While
maintaining these servers external to the Spark cluster potentially introduces
runtime costs associated with networking latencies this approach does allow
fast prototyping for ROSE-enabled Spark applications as no configuration
changes need to be made to an existing Spark cluster to get up and running.
To take advantage of an external OpenCPU server cluster when working with
ROSE simply pass an Array[String] of one or more server endpoints to the
analyze transformation. For example, identify an external cluster of 3
OpenCPU servers for use by your Spark application:
val ocpuCluster = sc.broadcast(Array("http://1.1.x.x/ocpu",
"http://1.1.y.y/ocpu",
"http://1.1.z.z/ocpu"))
val rResultRDD = rTaskRDD.analyze(ocpuCluster.value)
As shown, the array of server endpoints should be maintained as a
broadcast variable within your Spark application. Each OCPUTask within
your ROSE application will automatically be distributed for execution at random
across the OpenCPU server cluster.
See the LICENSE file for license rights and limitations (Apache License 2.0).