-
Notifications
You must be signed in to change notification settings - Fork 1
Atomer: Atomicity Violations Analyser
Atomer is a static analyser that detects atomicity violations. It is implemented as a module of Facebook Infer (an open-source static analysis framework). Atomer works on the level of sequences of function calls. It is based on the assumption that sequences executed atomically once should probably be executed always atomically. The implementation particularly targets C/C++/Java programs that use various lock mechanisms. It adapts the technique of contracts for concurrency.
The analysis is divided into two parts (phases of the analysis):
- Detection of sets of function calls that are likely to execute atomically (atomic sets).
- Detection of cases where the atomicity is broken (violations of the atomic sets).
These phases of the analysis and its workflow are illustrated in below:
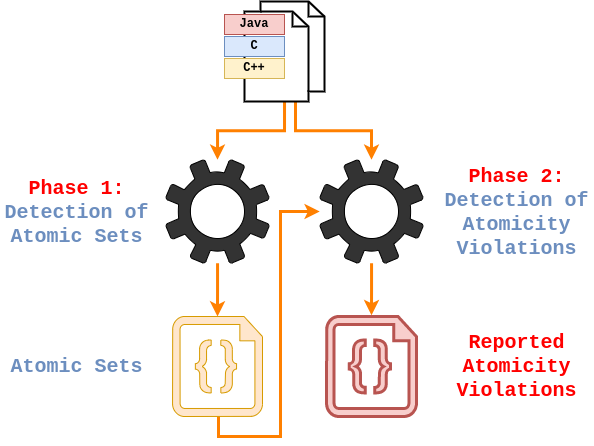
In the first phase of the analysis, functions index_of and set are stored into an atomic set because
they appear in an atomic block in function f.
void f(int *array)
{
pthread_mutex_lock(&lock);
int i = index_of(array, 42);
if (i >= 0) set(array, i, 3);
pthread_mutex_unlock(&lock);
}In the second phase, it is reported atomicity violation because the functions index_of and set from the atomic set are not called atomically in function g.
void g(int *array)
{
int i = index_of(array, 66);
if (i >= 0) set(array, i, 5);
}The implementation of Atomer can be found in the GitHub repository, which is a fork of the official repository of Facebook Infer, in branch atomicity-sets.
More details about Atomer can be found in the Related Papers section.
The whole installation process may be quite time-consuming.
At first, it is required to install Facebook Infer's dependencies and then to compile Facebook Infer. Here are the prerequisites to be able to compile Facebook Infer on Linux:
-
opam>=2.0.0 sqlitepkg-config-
Java(only needed for the Java analysis) -
gcc>=5.xorclang>=3.4(only needed for the C/Objective-C analysis) -
autoconf>=2.63 -
automake>=1.11.1 gmpmpfr-
CMake(only needed for the C/Objective-C analysis) -
Ninja(optional, if you wish to use sequential linking when building the C/Objective-C analysis)
See Atomer's Dockerfile for inspiration on how to install the dependencies on Linux. It includes the dependencies needed to build Infer on Debian 11 (Bullseye). Next, see the Atomer's GitHub Actions workflow file for inspiration on how to install the dependencies on Linux - Ubuntu 22.04 (Jammy Jellyfish) and macOS 12 (Monterey).
The installation of Facebook Infer can be done using the following commands:
# clone Facebook Infer using HTTPS...
git clone https://github.com/harmim/infer.git
# ..., or clone Facebook Infer using SSH
git clone git@github.com:harmim/infer.git
cd infer
# checkout Atomer branch
git checkout atomicity-sets
# build Facebook Infer
./build-infer.sh
make -j
# install Facebook Infer system-wide
sudo make install -j BUILD_MODE=opt
sudo chown -R "$USER" .The official installation manual can be found in INSTALL.md.
Facebook Infer now should be installed and executable by command infer. It is installed in infer/bin.
Alternatively, see how to run Atomer in Docker. The installation is much simpler and faster.
In general, to analyse a C/C++ program with Facebook Infer, it can be done using the following command (for a single file):
infer -- gcc -c file.cJava programs can be analysed using the following command:
infer -- javac file.javaAnother option is to analyse the entire project with Makefile using the following:
infer -- make <target>For advanced usage, see the Infer workflow. Many other build systems may be used, see Analyzing apps or projects.
Atomer is deactivated by default. The analysis have to be executed twice (it has two phases). Each phase runs with a different command line option. The first phase derives sets of functions that should be executed
atomically into file atomic-sets. The second phase then detects atomicity violations according to this file. So, the analysis can be triggered using the following commands:
infer capture -- gcc -c file.c
infer analyze --atomic-sets-only
infer analyze --atomicity-violations-onlyOr it can be triggered along with other analyses that Facebook Infer provides:
infer capture -- gcc -c file.c
infer analyze --atomic-sets
infer analyze --atomicity-violationsTo analyse Java programs, use the following commands:
infer capture -- javac file.java
infer analyze --atomic-sets-only
infer analyze --atomicity-violations-onlySee man infer or man infer-analyze.
-
--atomic-sets-file-append- Specify whether functions should be appended to the atomic sets file instead of overriding in the
atomic-setschecker. A default value isfalse.
- Specify whether functions should be appended to the atomic sets file instead of overriding in the
-
--atomic-sets-functions-depth-limit int- Specify the maximum depth in the hierarchy of function calls to which function calls will be considered during the
atomic-setschecker analysis. A default value is10.
- Specify the maximum depth in the hierarchy of function calls to which function calls will be considered during the
-
--atomic-sets-ignore-single-atomic-calls- Specify whether function calls that appear just once in critical sections should be ignored in the
atomic-setschecker. A default value isfalse.
- Specify whether function calls that appear just once in critical sections should be ignored in the
-
--atomic-sets-locked-functions-limit int- Specify the maximum number of function calls that could appear in a critical section in the
atomic-setschecker. Critical sections with more function calls will be ignored. A default value is20.
- Specify the maximum number of function calls that could appear in a critical section in the
-
--atomic-sets-widen-limit int- Specify the maximum number of iterations in a widening operator in the
atomic-setschecker. A default value is5.
- Specify the maximum number of iterations in a widening operator in the
-
--atomicity-allowed-classes path- Specify a file with class names (one class name per a line; considered as a regexp if the line starts with
Rfollowed by a whitespace, an exact match otherwise) whose method calls should be allowed during the analysis ofatomic-setsandatomicity-violationscheckers. Other calls will be ignored.
- Specify a file with class names (one class name per a line; considered as a regexp if the line starts with
-
--atomicity-allowed-function-analyses path- Specify a file with function names (one function name per a line; considered as a regexp if the line starts with
Rfollowed by a whitespace, an exact match otherwise) whose analysis should be allowed during the analysis ofatomic-setsandatomicity-violationscheckers. Other functions will be ignored.
- Specify a file with function names (one function name per a line; considered as a regexp if the line starts with
-
--atomicity-allowed-function-calls path- Specify a file with function names (one function name per a line; considered as a regexp if the line starts with
Rfollowed by a whitespace, an exact match otherwise) whose calls should be allowed during the analysis ofatomic-setsandatomicity-violationscheckers. Other functions will be ignored.
- Specify a file with function names (one function name per a line; considered as a regexp if the line starts with
-
--atomicity-ignored-function-analyses path- Specify a file with function names (one function name per a line; considered as a regexp if the line starts with
Rfollowed by a whitespace, an exact match otherwise) whose analysis should be ignored during the analysis ofatomic-setsandatomicity-violationscheckers.
- Specify a file with function names (one function name per a line; considered as a regexp if the line starts with
-
--atomicity-ignored-function-calls path- Specify a file with function names (one function name per a line; considered as a regexp if the line starts with
Rfollowed by a whitespace, an exact match otherwise) whose calls should be ignored during the analysis ofatomic-setsandatomicity-violationscheckers.
- Specify a file with function names (one function name per a line; considered as a regexp if the line starts with
-
--atomicity-lock-level-limit int- Specify the maximum expected level of ownership over the same lock object. An over-approximation of the number of times the lock has been acquired. A default value is
5.
- Specify the maximum expected level of ownership over the same lock object. An over-approximation of the number of times the lock has been acquired. A default value is
-
--atomicity-violations-ignore-local-calls- Specify whether function calls on local objects should be ignored in the
atomicity-violationschecker. A default value istrue.
- Specify whether function calls on local objects should be ignored in the
-
--atomicity-violations-widen-limit int- Specify the maximum number of iterations in a widening operator in the
atomicity-violationschecker. A default value is10.
- Specify the maximum number of iterations in a widening operator in the
Atomer prints all detected atomicity violations to the standard output and to files infer-out/report.txt, infer-out/report.json.
$ infer run --atomic-sets-only -- gcc -c main.cpp
Capturing in make/cc mode...
Found 1 source file to analyze in /.../infer-out
The detection of atomic sets produced an output into file '/.../atomic-sets'.
1/1 [################################################################################] 100% 127ms
No issues found
$ cat atomic-sets
Test::test_std_lock: {Test::f1, Test::f2} {Test::f1, Test::f2, Test::f3}
Test::test_lock_guard: {Test::f1, Test::f2, Test::f3} {Test::f3}
Test::test_std_mutex: {Test::f1, Test::f2} {Test::f1, Test::f2, Test::f3} {Test::f3}
Test::test_unique_lock: {Test::f2, Test::f3} {Test::f3}
# Number of (analysed functions; atomic sets; atomic functions): (4; 9; 18)
$ infer analyze --atomicity-violations-only
Found 1 source file to analyze in /.../infer-out
1/1 [################################################################################] 100% 24.729ms
main.cpp:79: error: Atomicity Violation
Atomicity Violation! - Functions 'Test::f1' and 'Test::f2' should be called atomically.
77. void test_violations(void)
78. {
79. f1(); f2(); // (f1, f2)
^
80. }
81. };
Found 1 issue
Issue Type(ISSUED_TYPE_ID): #
Atomicity Violation(ATOMICITY_VIOLATION): 1
If you have any questions, do not hesitate to contact the tool/method authors:
- Harmim, D. Advanced Static Analysis of Atomicity in Concurrent Programs through Facebook Infer. Brno, CZ, 2021. Master's thesis. Brno University of Technology, Faculty of Information Technology. Department of Intelligent Systems. Supervisor Vojnar, T.
- Harmim, D. Advanced Static Analysis of Atomicity in Concurrent Programs through Facebook Infer. In: Proceedings of the Excel@FIT'21. Brno, CZ: Brno University of Technology, Faculty of Information Technology, 2021.
- Harmim, D. Static Analysis Using Facebook Infer to Find Atomicity Violations. Brno, CZ, 2019. Bachelor's thesis. Brno University of Technology, Faculty of Information Technology. Department of Intelligent Systems. Supervisor Vojnar, T.
- Harmim, D.; Marcin, V.; Pavela, O. Scalable Static Analysis Using Facebook Infer. In: Proceedings of the Excel@FIT'19. Brno, CZ: Brno University of Technology, Faculty of Information Technology, 2019.
- Dias, R. J.; Ferreira, C.; Fiedor, J.; Lourenço, J. M.; Smrčka, A.; Sousa, D. G.; Vojnar, T. Verifying Concurrent Programs Using Contracts. In: 10th IEEE International Conference on Software Testing, Verification and Validation. Tokyo, Japan: IEEE Computer Society, Los Alamitos, CA, USA, March 2017, p. 196–206. ICST'17. DOI: 10.1109/ICST.2017.25. ISBN 9781509060313.
Atomer is MIT-licensed.
- The initial version
1.0.0of Atomer created within the H2020 ECSEL project AQUAS is available inatomer-v1.0.tgz. There is also GitHub releasev1.0.0, which also contains binaries. - The second major version
2.0.0of Atomer created within the H2020 ECSEL projects Arrowhead Tools and VALU3S is available in GitHub releasev2.0.0. It contains the source code as well as binaries. - The current version of Atomer is in the GitHub repository.
Development of this tool has been supported by the Arrowhead Tools project. This project has received funding from the Electronic Component Systems for European Leadership Joint Undertaking under grant agreement No. 826452. This Joint Undertaking receives support from the European Union's Horizon 2020 research and innovation programme and multiple EU countries, including the Czech Republic.
Development of this tool has been supported by the AQUAS project (Aggregated Quality Assurance for Systems). This project has received funding from the Electronic Component Systems for European Leadership Joint Undertaking under grant agreement No. 737475. This Joint Undertaking receives support from the European Union's Horizon 2020 research and innovation programme and Spain, France, United Kingdom, Austria, Italy, Czech Republic, Germany.
This tool as well as the information provided on this web page reflects only the author's view and ECSEL JU is not responsible for any use that may be made of the information it contains.


This project is co-funded by the European Union.