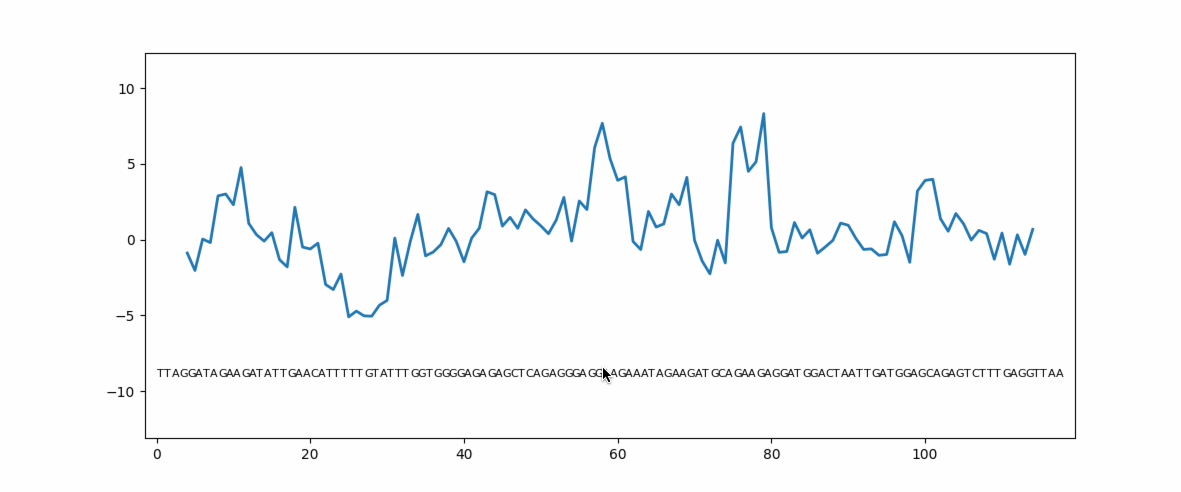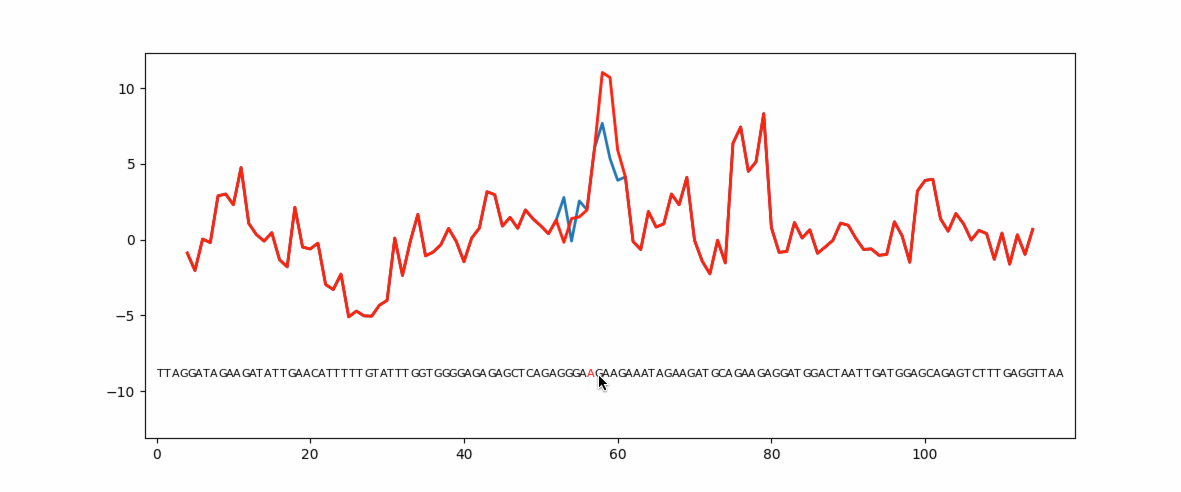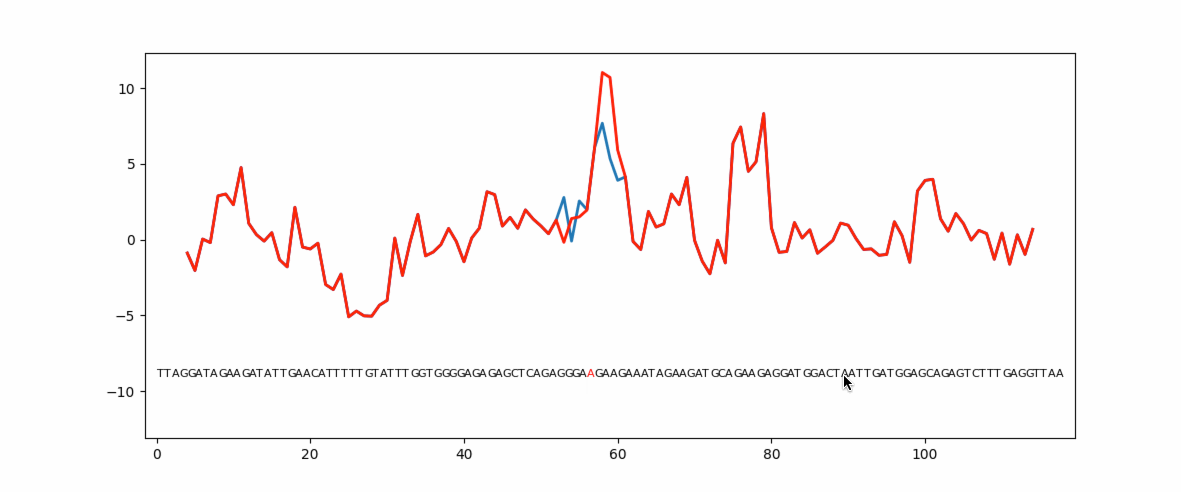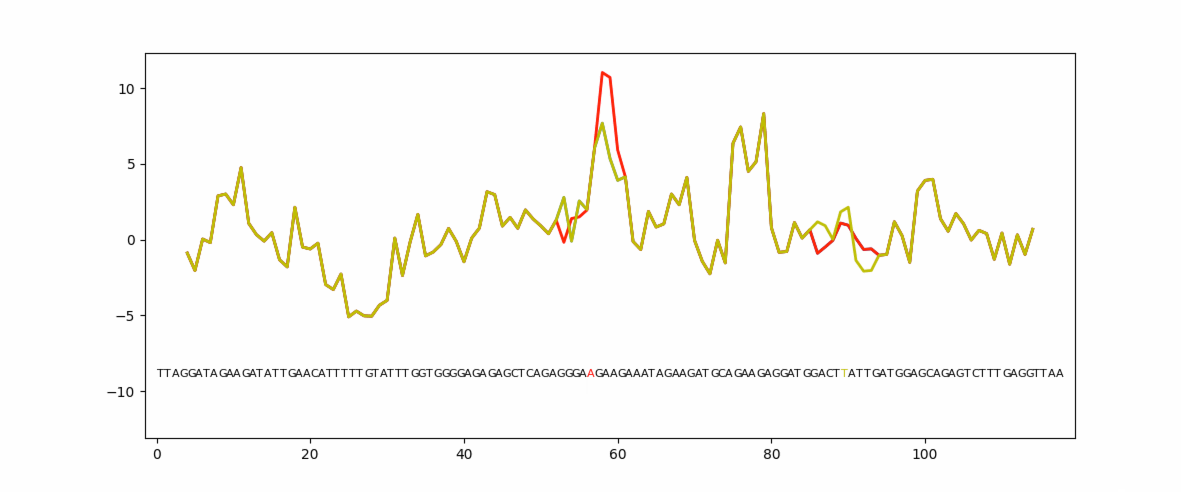
Eukaryotic genes have coding regions (exons) interrupted by non-coding introns. Exons must be selected and spliced together and introns removed. This process is governed by numerous short exonic sequences called splicing enhancers (ESE) or splicing silencers (ESS). The balance between ESE and ESS determines whether an exons is selected or not. Unsurprisingly, mutations in ESE or ESS sequences can cause human disease due to loss or gain of the exon. Mutation planner is a Python package that can help biologists to predict the loss or gain of ESE or ESS sequences.
pip install git+https://github.com/cssd2019/mutationplanner.gitimport mutationplanner as mp
octamers_file_path = "./data/octamers.txt"
fasta_file_path = "./data/test.fasta"
fasta_record_id = 'wt'
strategy = 2
# call to the main function of mutationplanner
mp.mutation_analyser(octamers_file_path, fasta_file_path, fasta_record_id, strategy)If you encounter a clear bug, please file a minimal reproducible example on github.
MIT