Tool to download / merge individual RNASeq files from the GDC Portal into a matrices identified by TCGA barcode.
Description:
The gdc-rnaseq-tool performs the following:
- Downloads RNA-Seq / miRNA-Seq data files using a GDC manifest file
- Unzips the files into separate folders identified by experimental strategy and bioinformatics workflow
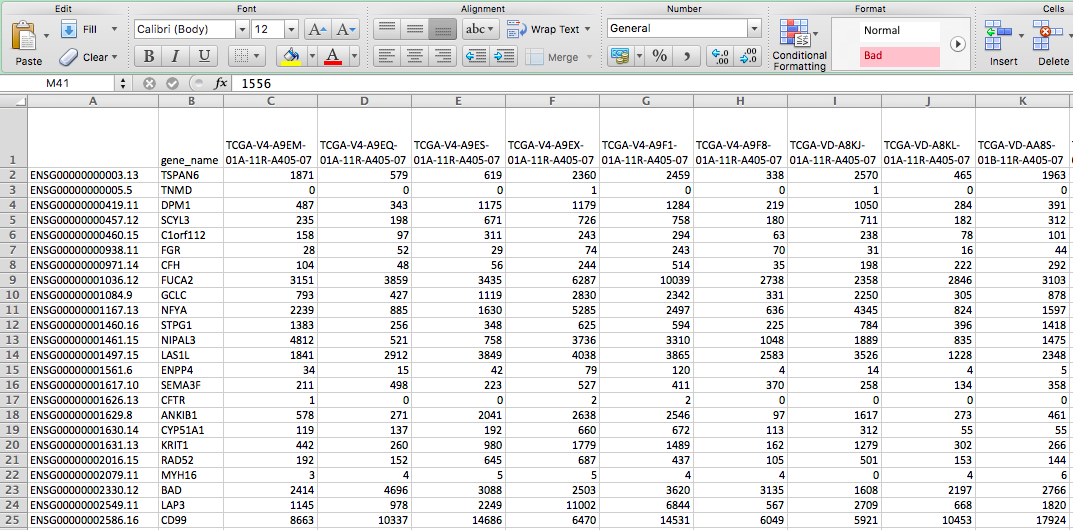
- Merges the files into separate matrix files identified in the table below
The script will ignore any files in the manifest file that are not Transcriptome Profiling files generated from the GDC RNA-Seq / miRNA-Seq bioinformatics pipelines located on the GDC Main Portal:
RNA-Seq Bioinformatics Pipeline Documentation
miRNA-Seq Bioinformatics Pipeline Documentation
Inputs and Outputs:
| I/O | File |
|---|---|
| Input | GDC Manifest File |
| Output | Merged_Counts.tsv (HTSeq - Counts) |
| Merged_FPKM.tsv (HTSeq - FPKM) | |
| Merged_FPKM-UQ.tsv (HTSeq - FPKM-UQ) | |
| Merged_miRNA_Counts.tsv | |
| Merged_miRNA_rpmm.tsv |
Requirements:
- Python 3+
- pandas ( https://pandas.pydata.org/pandas-docs/stable/install.html ):
pip3 install pandas - requests (http://python-requests.org):
pip3 install requests
Quick Start:
- Download
gdc-rnaseq-tool.pypython script - Download manifest containing RNA/miRNA expression files from https://portal.gdc.cancer.gov/
python3 gdc-rnaseq-tool.py <manifest_file>
Optional: Add --hugo to the command to include the HUGO gene symbol as a separate column.
python3 gdc-rnaseq-tool.py <manifest_file> --hugo
The GDC RNASeq tool produces matrices of merged RNA/MiRNA expression data given a manifest file.
Usage: python3 gdc-rnaseq-tool.py <manifest_file>
Notes:
- A test manifest is provided for troubleshooting:
python3 gdc-rnaseq-tool.py Test_Manifest.txt - Files are by default downloaded to the same folder as the manifest file that was provided
Release Notes:
Version 1.0: Feb 8, 2018
- Initial release
Known Issues: N/A