-
Notifications
You must be signed in to change notification settings - Fork 0
Initial Manuscript: Analysis and Evaluation Plan
Computational phenotyping (CP) leverages predefined sets of clinical concepts (e.g., diagnoses, medications, procedures, and laboratory test codes) to identify patients with and without a condition. CP approaches have great potential to aid in diagnosis, prognosis, therapeutic decision-making, and identification of mechanisms or novel biomarkers.
Existing methods face three unsolved barriers:
- CP definitions may have limited generalizability because they are tailored to specific source vocabularies or hospital systems.
- CP definitions may lack translational relevance because they primarily rely on clinical data requiring additional mapping to incorporate, for example, molecular or physiologic data.
- CP definitions may lack scalability because the current process for creating definitions is a time-consuming, iterative process requiring both domain expertise and external validation.
How can we solve these problems?
PheKnowVec is a novel method for deriving, implementing, and validating CPs that addresses these barriers by:
- The mapping of source vocabularies to standardized clinical terminology concepts, like those in the Observational Medical Outcomes Partnership (OMOP) common data model.
- The mapping of standardized clinical terminology concepts to linked open data, such as biomedical ontologies, has been shown to significantly improve the process of integrating and incorporating sources of non-clinical data.
- Embedding methods, which convert large complex heterogeneous data into scalable compressed vectors without semantic information loss, have successfully solved a wide range of problems in the biomedical domain.
We define the following terms, which will be used throughout this section:
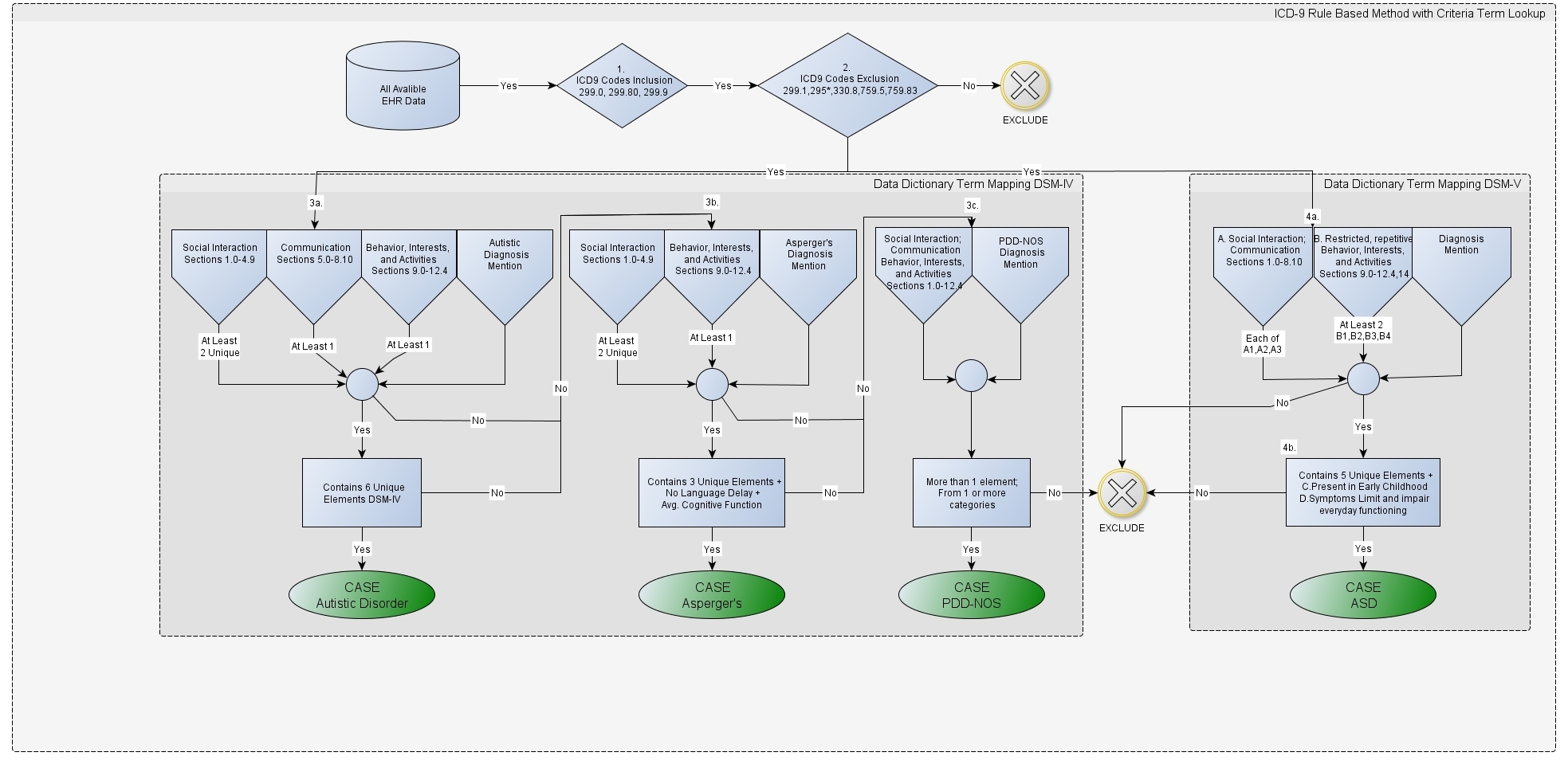
- Phenotype Definitions: A word document or a PDF that describes the clinical logic utilized to identify patients that have or do not have a specific disease. An example of what this logic looks like is shown in Figure below, which illustrates the PheKB Austism phenotype definition.
- Clinical Code Sets: Clinical codes (i.e. ICD9, SNOMED, RxNorm), which are compiled by the authors of the phenotype. These code sets are the product or output of following the logic described in the phenotype definition.
- Ontology code set: derived by mapping codes from the OMOP standardized terminologies to codes in a Open Biomedical Ontology (OBO).
The experiments described below will be evaluated using two independent datasets:
- The first dataset contains pediatric data and was extracted from a de-identified database built using data from the Children’s Hospital Colorado (CHCO). The CHCO De-ID data conforms to the structure defined by the Pediatric Learning Health System OMOP common data model version 5.
- The second dataset contains adult intensive care data built using data from the MIMIC III database that has also been standardized to the OMOP common data model version 5.3 (MIMICIII-OMOP).
All diagnoses, medications, laboratory tests, and procedures in the current build of each dataset will be considered for analysis. All patients whose record contained at least 1 code in at least 1 of the defined code sets will be included in the analysis. Use of this data was approved by the Colorado Multiple Institutional Review Board (protocol # 15-0445).
We will use all phenotypes appropriate for implementation in pediatric and adult populations from the eMERGE network's Phenotype KnowledgeBase (n=9). Table 1 provides an overview of the clinical domains and source vocabularies that are used in each of the phenotypes.
Additional information on the phenotypes listed below can be found within these documents:
TABLE 1. The eMERGE phenotypes selected for use in the PheKnowVec experiments.
| Phenotypes | Diagnoses | Medications | Lab Tests | Procedures | Problem Lists | Diagnoses Vocab | Medications Vocab | Lab Tests Vocab | Procedures Vocab | Problem List Vocab | NLP Required | Phenotype Definition | Case Definition | Control Definition |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ADHD | TRUE | TRUE | FALSE | FALSE | FALSE | ICD9-CM | String | --- | --- | --- | FALSE | FALSE | TRUE | TRUE |
| Appendicitis | TRUE | TRUE | FALSE | TRUE | FALSE | ICD9-CM | String | --- | CPT | --- | FALSE | TRUE | TRUE | TRUE |
| Crohn's Disease | TRUE | TRUE | FALSE | FALSE | FALSE | ICD9-CM | String | --- | --- | --- | FALSE | FALSE | TRUE | TRUE |
| Hypothyroidism | TRUE | TRUE | TRUE | TRUE | FALSE | ICD9-CM | String | String | CPT | --- | FALSE | TRUE | TRUE | TRUE |
| Peanut allergy | TRUE | FALSE | TRUE | TRUE | FALSE | String | --- | String | String | --- | FALSE | FALSE | TRUE | FALSE |
| Sickle Cell Disease | TRUE | FALSE | FALSE | FALSE | FALSE | ICD9-CM | --- | --- | --- | --- | FALSE | TRUE | TRUE | FALSE |
| Sleep Apnea | TRUE | FALSE | FALSE | FALSE | FALSE | ICD9-CM | --- | --- | --- | --- | FALSE | FALSE | TRUE | FALSE |
| Steroid-Induced Osteonecrosis | TRUE | TRUE | FALSE | TRUE | FALSE | ICD9-CM | String | --- | CPT | --- | FALSE | TRUE | TRUE | TRUE |
| Systemic Lupus Erythematosus | TRUE | TRUE | TRUE | FALSE | FALSE | ICD9-CM | String | String | --- | --- | FALSE | FALSE | TRUE | TRUE |
Clinical Code Sets: When a clinical terminology or vocabulary code is provided, they will be mapped to the following standard terminologies:
- Diagnoses and Problem Lists: SNOMED CT
- Drugs: RxNorm
- Measurements: LOINC
- Procedures: CPT4, HCPCS, ICD9Proc, CDT, ICD10PCS, OPCS4, and SNOMED CT
In the absence of being provided a code from a clinical terminology or vocabulary, the phenotype author will provide a string or descriptive phrase (i.e. "adderall", "history of appendicitis"). In these situations, the input or source string must then be mapped to a code in a clinical terminology or vocabulary (e.g. SNOMED CT, RxNorm, LOINC). While this may seem like a trivial task, the amount of potential matches between a source string and an existing concept in a clinical terminology or vocabulary can be immense. For example, the table below provides two different ways that one could map the source strings "adderall" and “appendicitis” to a clinical code.
| Source String | Results | Mapped Result Examples |
|---|---|---|
| adderall | 356 | - SPL: 2ea20fcc-3a59-4b9b-9b90-9da0d2bf219d - Adderall- RxNorm: 84815 - Adderall- SNOMED: 15846811000001101 - Adderall XR 30mg capsules (Imported (United States))- NDC: 54868367402 - Amphetamine aspartate 2.5 MG / Amphetamine Sulfate 2.5 MG / Dextroamphetamine saccharate 2.5 MG / Dextroamphetamine Sulfate 2.5 MG Oral Tablet [Adderall] |
| appendicitis | 154 | - CIEL: 121518 - Appendicitis- CIEL: 145490 - Chronic Appendicitis- SNOMED: 74400008 - Appendicitis- SNOMED: 286967008 - Acute perforated appendicitis- MeDRA: 10003011 - Appendicitis- MedDRA: 10066023 - Necrotising appendicitis |
As you can see from the table above, there are many different ways that a source string can be mapped to a SV code. The potential downstream impact of these different mappings on clinical diagnostics is not fully understood. To choose the best approach, we must first understand what all of the different options are and how/why they differ. This is the goal of the current verification task. For more information on how we have mapped and verified phenotype source strings to SV codes, please see the Code Sets tab of the Wiki.
Ontology Code Sets: The following mappings between ST clinical code sets and OBO ontology code sets (see the BioLater project for more information) were developed, by clinical domain:
- Diagnoses and Problem Lists: Over 29,000 unique ST clinical codes (SNOMED CT) were mapped to codes in the Human Phenotype Ontology and the Human Disease Ontology. We verified 1,000 manually mapped or constructed mappings with a clinician who had over 5 years of professional medical coding experience.
- Drugs: Over 8,000 unique ST clinical codes (RxNORM) were mapped to codes in the Chemical Entities of Biological Interest Ontology, the Vaccine Ontology, the Protein Ontology, and the NCBITaxon. We verified 25% of the ingredients that were manually mapped or constructed with a professional pharmacist.
- Measurements: Over 2,500 unique ST clinical codes (LOINC) were manually mapped to codes in the Human Phenotype Ontology. We verified 1,500 of the mappings with a professional ontologist in addition, a subset of 270 mappings were manually verified by three clinicians.
- Procedures: Over 14,000 ST clinical codes (CPT4, HCPCS, ICD9Proc, CDT, ICD10PCS, OPCS4, and SNOMED) were mapped to codes in the National Cancer Institute Thesaurus.
Generating Code Sets
Code sets were created for condition, medication, laboratory, and procedure concepts using a similar approach as Hripcsak et al., 2018. Queries to generate code sets can be found here. There are several important definitions to keep in mind when interpreting the table below:
- Knowledge Engineered (KE): The set of vocabulary codes were vetted by a domain expert
- No Descendants: The vocabulary code without any child or descendant codes.x
- Children: The vocabulary code with its immediate children only
- Descendants: The vocabulary code with all of its descendants
For example, the code for "Down Syndrome" has the following 9 children and 18 descendants:
| Codes | Child | Descendant |
|---|---|---|
| Partial trisomy of chromosome 21 | X | X |
| Anomaly of chromosome pair 21 | X | |
| Fetus with complete trisomy 21 syndrome | X | |
| Transient abnormal myelopoiesis co-occurrent with Down syndrome | X | |
| Trisomy 21- meiotic nondisjunction | X | |
| Partial trisomy 21 in Down's syndrome | X | X |
| Ring chromosome 21 syndrome | X | X |
| Deletion of part of chromosome 21 | X | X |
| Myeloid leukaemia co-occurrent with Down syndrome | X | |
| 21q partial trisomy syndrome | X | X |
| Complete monosomy 21 syndrome | X | X |
| Translocation Down syndrome | X | X |
| Periodontitis co-occurrent with Down syndrome | X | |
| Dementia co-occurrent and due to Down syndrome | X | |
| Trisomy 21- mitotic nondisjunction mosaicism | X | X |
| 21q partial monosomy syndrome | X | |
| Complete trisomy 21 syndrome | X | X |
| 21q partial distal trisomy syndrome | X |
Given the definitions above, the following code sets will be constructed and compared:
TABLE 2. Code sets used in PheKnowVec experiments.
| Code Set | Match Type | Knowledge | Count | Description | No Descendants | Child | Descendants |
|---|---|---|---|---|---|---|---|
| Source Vocabularies | Exact | None | 3 | These code sets are comprised of all codes and strings provided by the eMERGE phenotype authors. When a string is provided, each single-word string is exact AND fuzzy matched to concepts in the OMOP concept table. | X | X | X |
| Synonyms | 3 | X | X | X | |||
| Domain Expert | 3 | X | X | X | |||
| Fuzzy | None | 3 | X | X | X | ||
| Synonyms | 3 | X | X | X | |||
| Domain Expert | 3 | X | X | X | |||
| Standard Terminologies | Exact | None | 3 | These code sets are comprised by using the OMOP concept_relationship table to map source vocabulary codes from the source vocabulary - exact and source vocabulary - fuzzy code sets to codes in specific standard terminologies. | X | X | X |
| Domain Expert | 3 | X | X | X | |||
| Fuzzy | None | 3 | X | X | X | ||
| Domain Expert | 3 | X | X | X | |||
| Open Biomedical Ontologies | Exact | None | 3 | These code sets are comprised by automatically mapping (i.e., using exact string matches and database cross-references) standard terminology codes to codes in an OBO. | X | X | X |
| Domain Expert | 3 | X | X | X | |||
| Manual | None | 3 | These code sets are comprised by manually mapping (i.e., using a _intersection_of()_, _union_of()_, or _disjoint_from()_ constructors) standard terminology codes to codes in an OBO. | X | X | X | |
| Domain Expert | 3 | X | X | X |
NOTE. As indicated in the OMOP documentation, it is not possible to find ancestors or descendants of deprecated or Source Concepts. This means that when a source_code is provided in the phenotype definition, we do not search for it's children/descendants. We do search for children/descendants when an input_string is provided because they could be exact- or fuzzy-matched to a standard_concept, which are linked to ancestors/descendants.
To demonstrate the generalizability, translatability, and scalability of PheKnowVec, we will perform several experiments drawing from the following four categories:
-
Phenotype Cohort Assignment: patients will be assigned to a cohorts in two ways:
- Having >=1 occurrence of 1 or more codes specified by the inclusion criteria in the phenotype definition (Phenotype Codes)
- Exception: If the phenotype has a control group, then the presence >=1 occurrence of 1 or more exclusion criteria in the phenotype definition is also used.
- Having the codes and meeting logic specified in the phenotype definition (Phenotype Definitions)
- Having >=1 occurrence of 1 or more codes specified by the inclusion criteria in the phenotype definition (Phenotype Codes)
- Cohort Group: cases vs. controls
- Code Sets: each of the code sets defined in Table 2
- Clinical Data Types: Only Conditions vs. All Clinical Domains (conditions, medications, laboratory tests, and procedures)
The generalizability and translatability experiments will be performed using the CHCO OMOP DeID and the MIMICIII-OMOP data. The MIMICIII-OMOP data will only be used in the scalability task to validate the representations built from CHCO OMOP DeID.
For each phenotype, we will examine what information is gained and/or lost when deriving pediatric and adult patient cohorts using different the clinical code sets defined in Table 2. For all comparisons, the Source Vocabulary - Exact None code set will be used as the gold standard.
For each phenotype, the comparisons listed in Table 3 (shown below) will be performed and evaluated using:
- False Negative and False Positive Error Rate: the number of incorrectly included (false positive or FP) or missed (false negative or FN) patients using each standard terminology clinical code set versus the source vocabulary clinical code set (i.e. gold standard patient cohort) for cases and controls.
- Matthews Correlation Coefficient (MCC): The MCC ranges from -1 to +1 where -1 indicates the observed and expected binary classifications disagree, 0 indicates the agreement is no better than random chance and +1 indicates the observed and predicted binary classifications completely agree. This measure takes advantage of all four cells of the confusion matrix, which makes it well-suited for unbalanced classes.
- Cohort Verification: randomly select a small subset of FP and FN patients assigned to one or two phenotypes for review by a clinician.

TABLE 3. Comparisons to evaluate the generalizability of mapping clinical code sets.
| Phenotype Cohort Assignment | Cases | Controls | Code Sets | Clinical Data Types |
|---|---|---|---|---|
| Phenotype Codes | X | X | Source Vocabularies | Only Condition |
| Phenotype Codes | X | X | Source Vocabularies | All Clinical Domains |
| Phenotype Definitions | X | X | Source Vocabularies | Only Condition |
| Phenotype Definitions | X | X | Source Vocabularies | All Clinical Domains |
| Phenotype Codes | X | X | Standard Terminologies | Only Condition |
| Phenotype Codes | X | X | Standard Terminologies | All Clinical Domains |
| Phenotype Definitions | X | X | Standard Terminologies | Only Condition |
| Phenotype Definitions | X | X | Standard Terminologies | All Clinical Domains |
For each phenotype, we will examine what information is gained and/or lost when deriving pediatric and adult patient cohorts when ST clinical code sets are mapped to ontology code sets (Figure). For all comparisons, the Source Vocabulary - Exact None code set will be used as the gold standard.
For each phenotype, the comparisons listed in Table 4 (shown below) will be performed and evaluated using:
- False Negative and False Positive Error Rate: the number of incorrectly included (false positive or FP) or missed (false negative or FN) patients using each standard terminology clinical code set versus the source vocabulary clinical code set (i.e. gold standard patient cohort) for cases and controls.
- Matthews Correlation Coefficient (MCC): The MCC ranges from -1 to +1 where -1 indicates the observed and expected binary classifications disagree, 0 indicates the agreement is no better than random chance and +1 indicates the observed and predicted binary classifications completely agree. This measure takes advantage of all four cells of the confusion matrix, which makes it well-suited for unbalanced classes.
- Cohort Verification: randomly select a small subset of FP and FN patients assigned to one or two phenotypes for review by a clinician.
- To illustrate the translatability of this approach, we will take 1 or 2 of the phenotypes and extend their ontology code sets to include additional open data sources that are already manually annotated to the ontologies (e.g. DOID and HPO contain hand-annotated gene list mappings).
- We will perform basic clustering within each phenotype to derive sub-groups. Identify the most important codes within each cluster and get a domain expert to help with interpreting the results.

TABLE 4. Comparisons to evaluate the translatability of mapping clinical code sets to ontology code sets.
| Phenotype Cohort Assignment | Cases | Controls | Code Sets | Clinical Data Types |
|---|---|---|---|---|
| Phenotype Codes | X | X | Standard Terminologies | All Clinical Domains |
| Phenotype Definitions | X | X | Standard Terminologies | All Clinical Domains |
| Phenotype Codes | X | X | Open Biomedical Ontologies | All Clinical Domains |
| Phenotype Definitions | X | X | Open Biomedical Ontologies | All Clinical Domains |
For each phenotype, we will create patient-level embeddings for each of the cohorts that were derived using the clinical and ontology code sets from the translatability experiments (Figure). Two types of patient-level embeddings will be built. The first type of embedding will include only the clinical codes explicitly outlined by the phenotype definition. The second type of embedding will be built using all available data. For all comparisons, the best performing clinical and ontology code sets (without descendants) from the translatability experiments will be used as gold standards.
For each pediatric phenotype, the comparisons listed in Table 5 (shown below) will be performed and evaluated using the approaches described below:
-
Leave-One-Patient-Out (LOPO) Cross Validation with Logistic Regression and Youden Index Thresholding: within each phenotype, the cosine similarity between each patient and all other patient’s vectors will be calculated. The Youden Index is then used to convert the continuous cosine similarity score for each pairwise patient comparison into a cut-off that can be used for binary classification. This task will be performed for cases and controls, within each phenotype, as well as pooled so that we can determine how well the definition vectors identify the correct patients within each case and control group, across all phenotypes.
- Performance metrics for each case and control group by phenotype: accuracy, precision, recall, ROC curves and counts of TP for most similar 1, 5, 10, 25, 50, 75, and 100 patients.
-
Aggregated Case and Control Phenotype Definition Vectors: We will apply the same approach described above, but will compare all patients within each case and control group for each phenotype to the aggregated case and control phenotype definition vectors. The best performing code set phenotype definition vectors (1 clinical and 1 ontology code set) will be applied to the OMOP MIMIC data.
- Performance will be evaluated through domain expert-review of patient groups returned for each aggregated cohort group within each phenotype.

TABLE 5. Comparisons to evaluate the scalability of embedded clinical and ontology code sets.
| Phenotype Cohort Assignment | Cases | Controls | Code Sets | Clinical Data Types | Embedded Data |
|---|---|---|---|---|---|
| Phenotype Definitions | X | X | Standard Terminologies | All Clinical Domains | Only phenotype code data |
| Phenotype Definitions | X | X | Standard Terminologies | All Clinical Domains | All available data |
| Phenotype Definitions | X | X | Open Biomedical Ontologies | All Clinical Domains | Only phenotype code data |
| Phenotype Definitions | X | X | Open Biomedical Ontologies | All Clinical Domains | All available data |