Reproduction GIF below. On plate_list.html, user can pick any compressed gDNA plates and send them to the normalization.html page--but normalization can only be performed on compressed gDNA plates that have been quantified (via the parse_quantification.html page) . The normalization page prevents users from adding unquantified plates on it directly but does not catch the case where an unquantified plate is sent in via the querystring from plate_list.html. This means the user can go ahead and submit their normalization request to the back end and get a nasty python error.
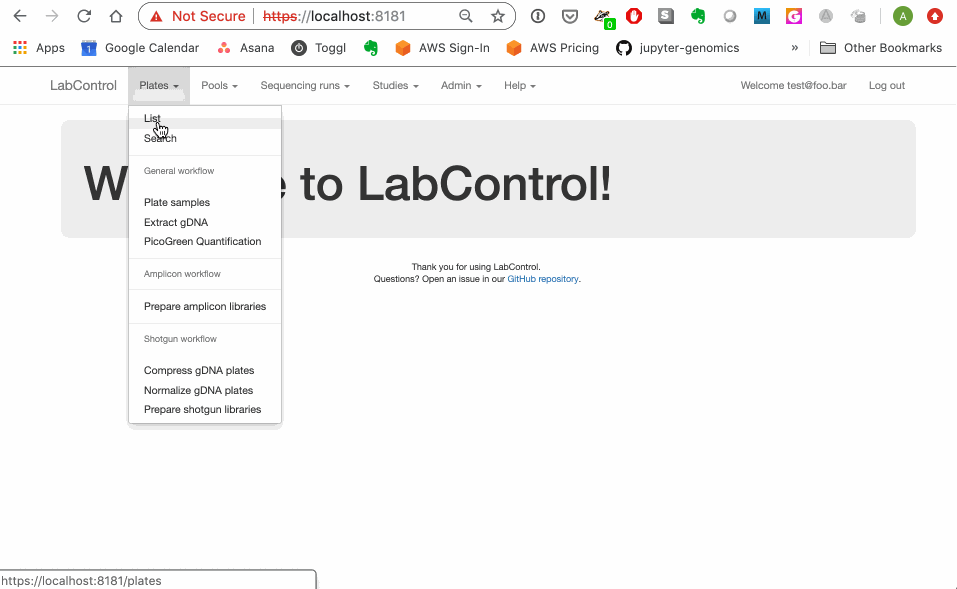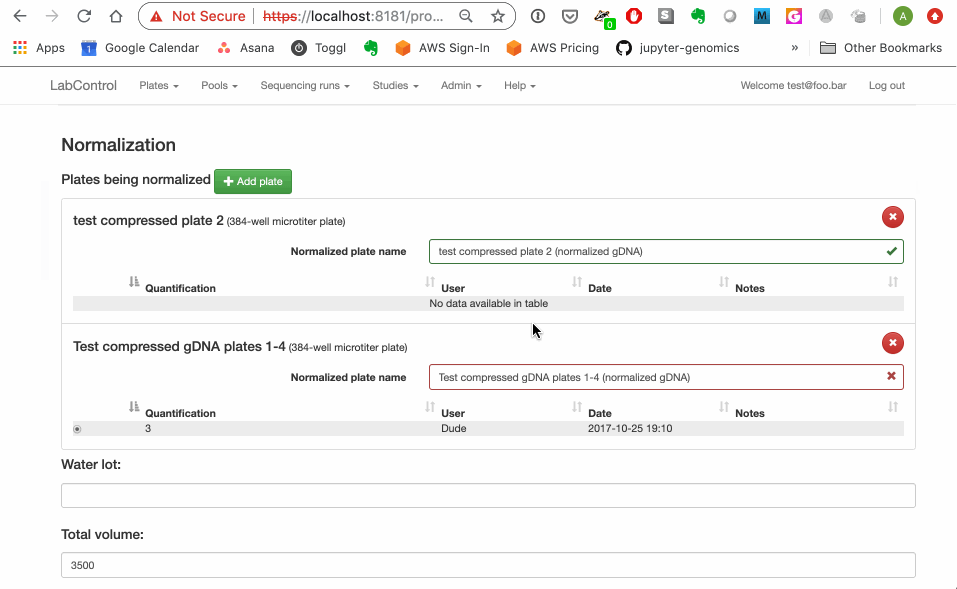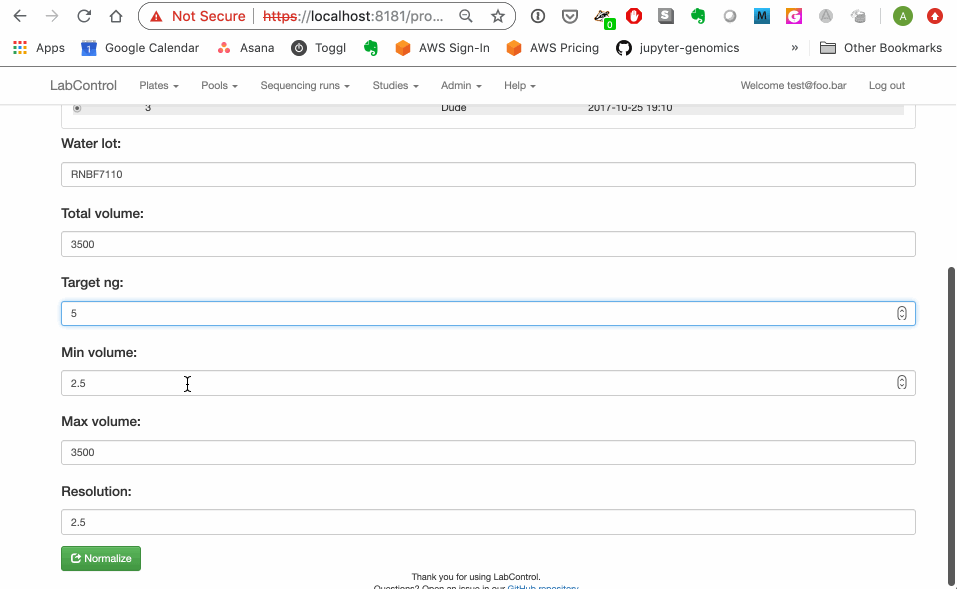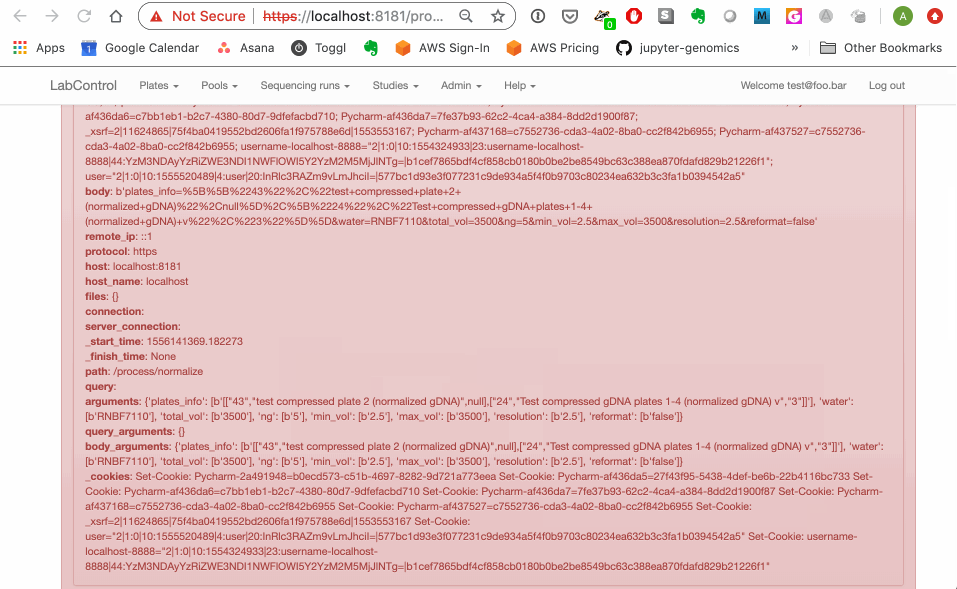
I class this as a relatively low priority because I don't think the lab actually uses the "checkboxes from plate list" route very often. It is actually a big pain to manage and I would like to suggest we at least consider the possibility of axing it ...