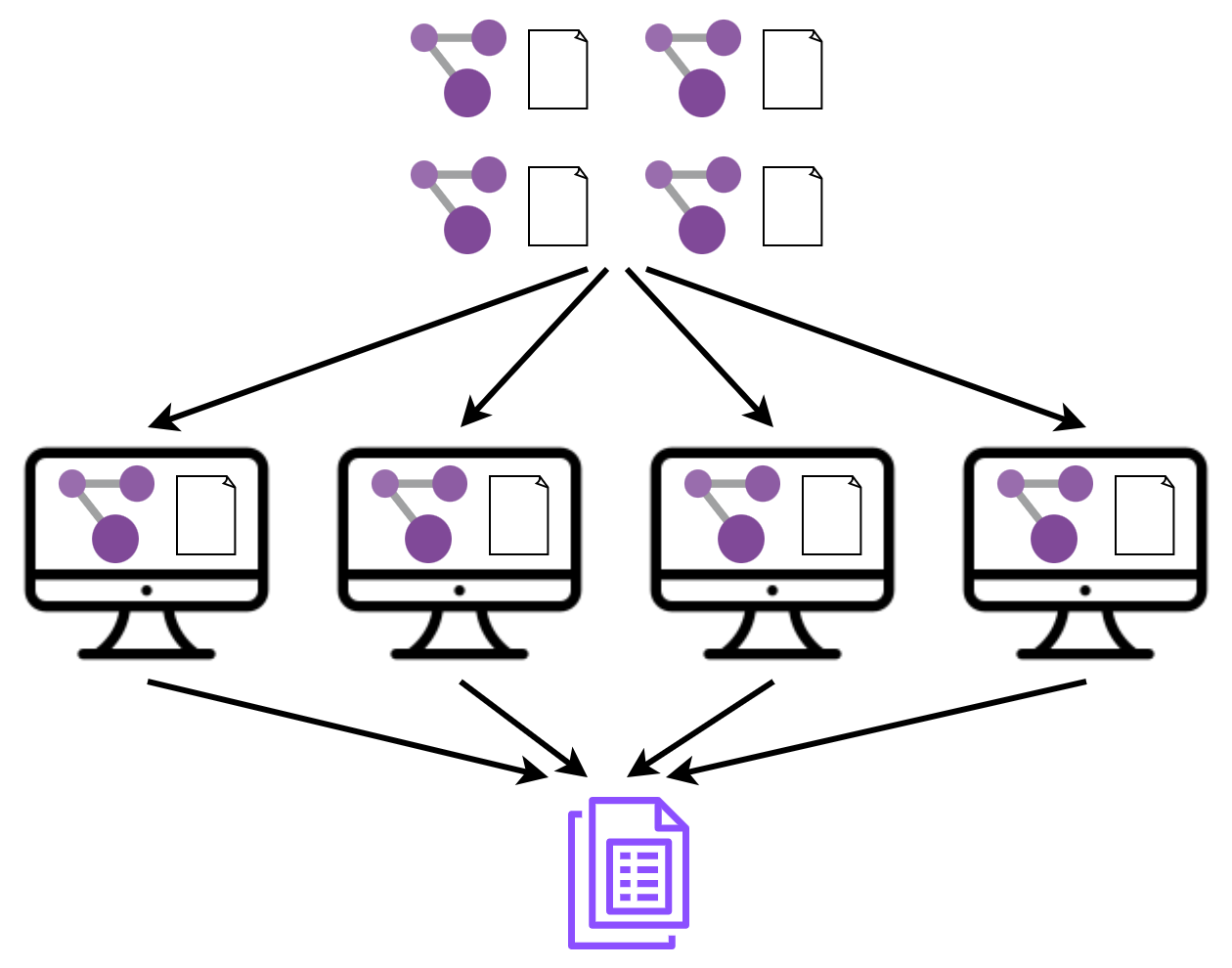
As part of the PaCER Hackathon 2023 I set out to learn and refresh my understanding of distributed computing and see if I can apply those concepts to distribute and bin multiple metagenomes. I also wanted to learn about the message passing interface (MPI). Here is the basic idea of my project.
I used two approaches.
- Array jobs
- MPI
Using the SLURM --array and passing a list of files to use for each job, I was able to distribute the binning jobs to different nodes. Check out the metacoag.slurm file for further details.
This was the challenging part. Rather than rewriting my code to distribute chunks of one dataset, I decided to distribute one dataset per node using MPI, do the binning, gather the results and do some statistics.
Make sure you have a working MPI implementation like MPICH or OpenMPI.
As my code was in python, I used the MPI python interface mpi4py which you can install as explained here. Make sure to match the compatible mpi4py version to the MPI version you have. I highly recommend staying away from prebuilt version and building from source.
I downloaded the source code of MetaCoAG from GitHub and installed it using pip locally.
git clone https://github.com/Vini2/MetaCoAG.git
cd MetaCoAG/
pip install -e .from mpi4py import MPI
def run_metacoag(folder_name, rank):
# bin dataset
# get nbins
return nbins
if __name__ == "__main__":
comm = MPI.COMM_WORLD
rank = comm.Get_rank()
size = comm.Get_size()
if rank == 0:
# List of input folders
input_folders = ["/path/to/test_dataset_1",
"/path/to/test_dataset_2"]
# Scatter the folders
for i in range(1, size):
comm.send(input_folders[i], dest=i)
# Receive the folder
if rank == 0:
received_data = input_folders[0]
else:
received_data = comm.recv(source=0)
# Process the received folder
results = run_metacoag(received_data, rank)
# Gather the number of bins from all processes
all_results = comm.gather(results, root=0)
if rank == 0:
# Process the gathered results
final_result = sum(all_results)
print("Total number of bins produced:", final_result)Basically, we have a set of folder paths in input_folders and we scatter them to nodes (done by comm.send). Then the nodes receive the data (done by comm.recv) and process it. Finally, the results are gathered (done by comm.gather and processed (here the total is calculated).
Change input_folders to your list of folders, load the relevant modules (e.g., MPI and mpi4py) and launch the SLURM script using,
sbatch mpi_metacoag.slurm
This is a crazy idea I came up with and might be totally useless. But I hope it might be useful for someone who wants to distribute files and run a command on multiple nodes using MPI.
I want to actually use MPI to distribute large graphs and do clustering on chunks. Fingers crossed! This was a great learning experience.