-
Notifications
You must be signed in to change notification settings - Fork 230
Description
Some time ago I wrote down a quick and rough implementation of the elliptical slice sampling algorithm in the Julia package EllipticalSliceSampling.jl (actually, recently I rewrote it almost completely but I haven't pushed the changes yet). Since it's a quite simple algorithm I thought it would be fun to implement an elliptical slice sampler for Turing.
I've managed to put together something that seems to work on a small and simple example. However, since I'm quite new to Turing and still don't understand all internals, I'm pretty sure my draft could be improved. Honestly, it took me quite some time to get a rough feeling for which types should be implemented and which methods should be overloaded (I think #895 (comment) was the most helpful resource I could find - thank you @cpfiffer for this write-up!). It was also surprisingly complicated to figure out how to sample from the prior and how to evaluate the log-likelihood (hopefully I actually managed to do it somewhat correctly in the end 😄 ).
I came up with the following implementation: https://gist.github.com/devmotion/6bab2561a9c340bac03c50b3fec73441#file-ess-jl Basically I tried to copy and adapt the MH implementation. However, I don't know if anything important is missing or wrong, or if some parts are not needed at all. I would be very happy if someone with a better understanding of Turing could comment on that! I'm also wondering if there is a way to avoid the repeated allocations caused by sampling from the prior? In the non-Turing implementation I used a cache to avoid these. One thing one would want to add is a check that the prior distribution is a uni- or multivariate normal distribution. Where would be the best place to do that? In the constructor of the sampler? Or in the implementation of assume?
For the simple model
@model gdemo(x) = begin
m ~ Normal(0, 1)
x ~ MvNormal(fill(m, length(x)), sqrt(σ²) * I)
endand 10 random observations (you can check the code) the sampler produced the following MCMC chain of 20000 samples:
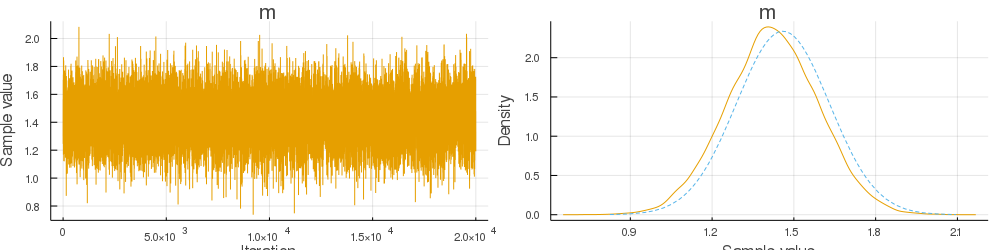
The comparison of the empirical distribution and the true posterior shows that the estimate of the mean (1.4210542073603993) is close to the true mean of the posterior (1.4597765432573337).
I'm happy to hear your comments and thoughts!
Edit: I reran the experiments since I missed that MvNormal assumes that a UniformScaling covariance is actually a matrix of standard deviations (to me that seems like a bug in Distributions).