A clear and interpretable baseline for predicting drug categories using patient features and a Decision Tree classifier. Designed to be interview-friendly, with emphasis on clarity, step-by-step decisions, and interpretability.
├── Drug_Prediction_DecisionTree_polished.ipynb
├── README.md # Project documentation
- Python, Jupyter Notebook
- pandas, NumPy — Data handling
- matplotlib, seaborn — Visualization
- scikit-learn — DecisionTreeClassifier, model evaluation
- EDA, Preprocessing, Model interpretation
This notebook demonstrates a complete Machine Learning workflow for predicting drug categories:
- Exploratory Data Analysis (EDA) – Inspect dataset distribution and patterns
- Preprocessing – Encoding categorical features, handling data types
- Model Training – Decision Tree Classifier
- Evaluation – Accuracy, interpretability, decision paths
- Features:
- Age: Age of the patient
- Sex: Male/Female
- Blood Pressure: Low / Normal / High
- Cholesterol: Normal / High
- Na_to_K ratio: Sodium-to-Potassium ratio in the blood
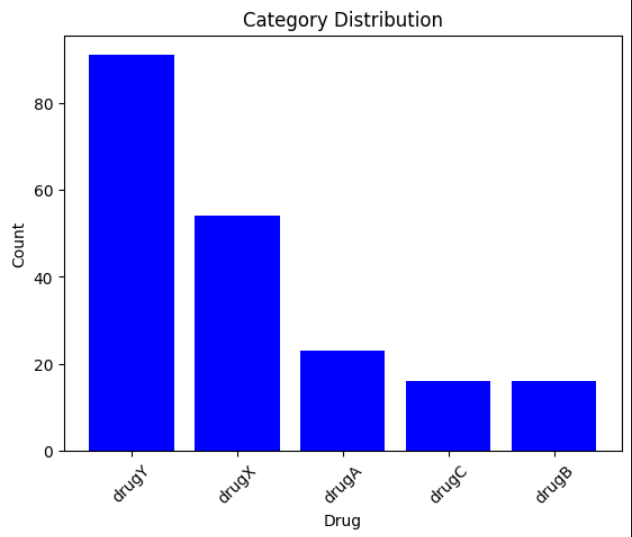
- Target: Drug type (DrugA, DrugB, DrugC, DrugX, DrugY)
- Size: 200 samples
- Source: UCI / educational dataset
- Clone or download this repository:
git clone https://github.com/Shamir-Havas/Drug-Prediction-Decision-Tree.git cd Drug-Prediction-Decision-Tree
Install dependencies:
bash Copy code pip install -r requirements.txt Open Jupyter Notebook and run the workflow:
bash Copy code jupyter notebook Drug_Prediction_DecisionTree_polished.ipynb Run all cells:
Kernel → Restart & Run All
📊 Results
🔹 Category Counts
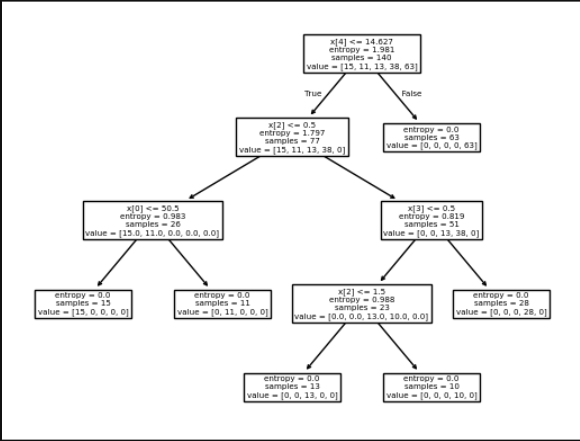
🔹 Decision Tree Visualization
🔹 Model Accuracy
🔍 Model Explainability
Decision Tree Visualization: Interpretable decision paths using plot_tree
Classification Report: Precision, recall, F1-score
🚀 Future Improvements
Hyperparameter tuning with GridSearchCV / RandomizedSearchCV
Cross-validation (e.g., Stratified K-Fold) for robustness
Try ensemble methods (Random Forest, XGBoost)
Domain-specific validation & feature engineering
📦 Requirements
pandas==2.0.3
numpy==1.25.2
matplotlib==3.7.2
seaborn==0.12.2
scikit-learn==1.3.0