-
Notifications
You must be signed in to change notification settings - Fork 154
Update BACs #563
New issue
Have a question about this project? Sign up for a free GitHub account to open an issue and contact its maintainers and the community.
By clicking “Sign up for GitHub”, you agree to our terms of service and privacy statement. We’ll occasionally send you account related emails.
Already on GitHub? Sign in to your account
Update BACs #563
Conversation
|
@kspieks Can you rebase this PR? Thanks |
|
Just rebased :) |
There was a problem hiding this comment.
Choose a reason for hiding this comment
The reason will be displayed to describe this comment to others. Learn more.
Thank you Kevin! Before this PR is merged. Can you please respond to my comments?
| 'N': -4.999999999999999, | ||
| 'O': -4.999999999999999, | ||
| 'S': -4.999999999999999 | ||
| }, |
There was a problem hiding this comment.
Choose a reason for hiding this comment
The reason will be displayed to describe this comment to others. Learn more.
No action is needed, this is an irrelevant comment to this PR. I feel like this is a bad fit since most parameters are just at the constraint boundary. Maybe someone needs to have a double-check on this. I don't see this happen to other Melius BACs
There was a problem hiding this comment.
Choose a reason for hiding this comment
The reason will be displayed to describe this comment to others. Learn more.
| units: kcal/mol | ||
| value: 48.724776954869256 | ||
| value: 48.479753141609 | ||
| class: ThermoData |
There was a problem hiding this comment.
Choose a reason for hiding this comment
The reason will be displayed to describe this comment to others. Learn more.
@kspieks Can you please comment on this? I do see a much larger difference before and after the change for ccsd(t)f12/ccpvdzf12 and ccpvtzf12 (O(0.1)) than wb97xd3/def2tzvp(O(0.01)). They are using the same un-scaled frequencies values, right? What I feel confused about is that the freq scaling factor for ccsd(t)f12 families is almost 1 (0.998 or likewise), and wb97xd3 is like 0.984. So ccsd(t)f12 should be less influenced by the mistake? Then why do we see a larger change?
There was a problem hiding this comment.
Choose a reason for hiding this comment
The reason will be displayed to describe this comment to others. Learn more.
I'm not sure. Both use the same frequency and scaling factor. Maybe I had accidentally used the ccsdtf12 scaling factor for the ccsdtf12 values last time, but I'm not sure. I definitely used the wb97xd3 freq scale factor of 0.984 this time since that is the LoT that the frequencies were calculated at. You can check the work in this notebook (which is identical to the BAC notebook we just merged in PR #501).
Peform BAC fitting-Copy1.ipynb.zip
There was a problem hiding this comment.
Choose a reason for hiding this comment
The reason will be displayed to describe this comment to others. Learn more.
Well, my point is the enthalpy is generated from the enthalpy of each mode + E0 + ZPE + AEC. Since in the original notebook, ZPE correction is considered, E0 + ZPE + AEC + enthalpy_translation + enthalpy_rotation should be the same, and the only difference is coming from enthalpy_vibration. No matter vibrational freq scale factor is considered or not, it shouldn't make huge impact for ccsd(t)f12 levels since their freq_scale is like 0.998, very close to 1. In my hypothesis, the difference before and after should be smaller than wb97xd levels (as their freq scale factors are more different from 1). However, I see some diff in ccsd(t)f12/cc-pvdz-f12 values are like 0.3 - 0.6 and some diffs in ccsd(t)-f12/cc-pvtz-f12 are even larger than 1, but diffs for wb97xd are like ~0.005. So I am confused about why such large difference happens.
You know, I just want to make sure the data used to fit BACs are correct. Since you have the data and the notebooks, can you help me check the enthalpy for 1-(Methylthio)butane (for example) obtained from your old and new notebook? Can you show me the term-wise values? energy from energy_log, freq_log.load_zero_point_energy() * zpe_scale_factor and conformer.get_enthalpy(298)? I appreciate your help for reviewing this PR.
There was a problem hiding this comment.
Choose a reason for hiding this comment
The reason will be displayed to describe this comment to others. Learn more.
I see your point and agree that it is helpful to go through one example molecule term by term. I agree that it's important to make sure the data used to fit the BACs are correct :) As expected, the frequency scaling factor, single point energy, and AEC are identical. It's only the ZPE that is different. However, the conformer.get_enthalpy(298) is slightly different as well since the new notebook from #501 correctly scales the frequencies which impacts the Cp that is integrated to get H298. For 1-(Methylthio)butane,
old notebook:
Used frequency scaling factor of: 0.984
sp energy: -1562740675.8574862 J/mol
ZPE: 419700.922752 J/mol
AEC: 1562191704.5336924 J/mol
sp energy + ZPE + AEC = conf E0 : -129270.40104198456 J/mol
H298: 25201.932158755808 J/mol
conf E0 + H298 = -24.87296101415601 kcal/mol, which matches the old value
new notebook:
Used frequency scaling factor of: 0.984
sp energy: -1562740675.8574862 J/mol
ZPE: 413906.23545562127 J/mol
AEC: 1562191704.5336924 J/mol
sp energy + ZPE + AEC = conf E0: -135065.0883383751 J/mol
H298: 25430.435136322696 J/mol
conf E0 + H298 = -26.2033109947544 kcal/mol, which matches the new value
Let me know if there are follow up questions. Happy to call briefly tomorrow if that's easier or if we should go through more examples.
There was a problem hiding this comment.
Choose a reason for hiding this comment
The reason will be displayed to describe this comment to others. Learn more.
Thank you so much, Kevin! This is really helpful. I don't understand why ZPEs are different, for both notebook you calculate the ZPE by freq_log.load_zero_point_energy() * zpe_scale_factor and zpe_scale_factor = freq_scale_factor / 1.014. I assume freq_log and freq_scale_factor are the same, then why ZPE is different? Once this is clear, I don't have further questions
There was a problem hiding this comment.
Choose a reason for hiding this comment
The reason will be displayed to describe this comment to others. Learn more.
In the original draft of the BAC notebook last year, there was some confusion about whether we should divide by 1.014 for all LoT or just for the g4mp2 data the Oscar and Duminda were working on. After doing more reading, we learned that we should always divide by 1.014 since this is an empirically derived value that relates frequency scaling factors to ZPE scaling factor to account for anharmonicity. This value is correctly hardcoded in Arkane and more details can be found in the 2010 publication by Alecu et al. The same frequency scaling factor was used in both notebooks. However, the difference in ZPE is because the first draft of the notebook did not divide by 1.014 while the updated version correctly does the division. Sorry for any confusion and thanks for taking the time to review this PR and check the values 🙂
There was a problem hiding this comment.
Choose a reason for hiding this comment
The reason will be displayed to describe this comment to others. Learn more.
Thanks again. When I looked at the latest notebook and the one you sent to me last year, they both have zpe_scale_factor = freq_scale_factor / 1.014, so I thought your value should get the same ZPE. I don't realize values for CCSD(T)-F12 are fitted using an even earlier version of the notebook.
OK, all set. Let me merge this PR in.
There was a problem hiding this comment.
Choose a reason for hiding this comment
The reason will be displayed to describe this comment to others. Learn more.
I had forced pushed the changes so yea it had erased the commit history that had incorrectly not divided by 1.014. Thanks again for your help in reviewing this PR
|
FYI: all log files and Arkane outputs related to these jobs have been copied to my account on our new greencloud sever at |
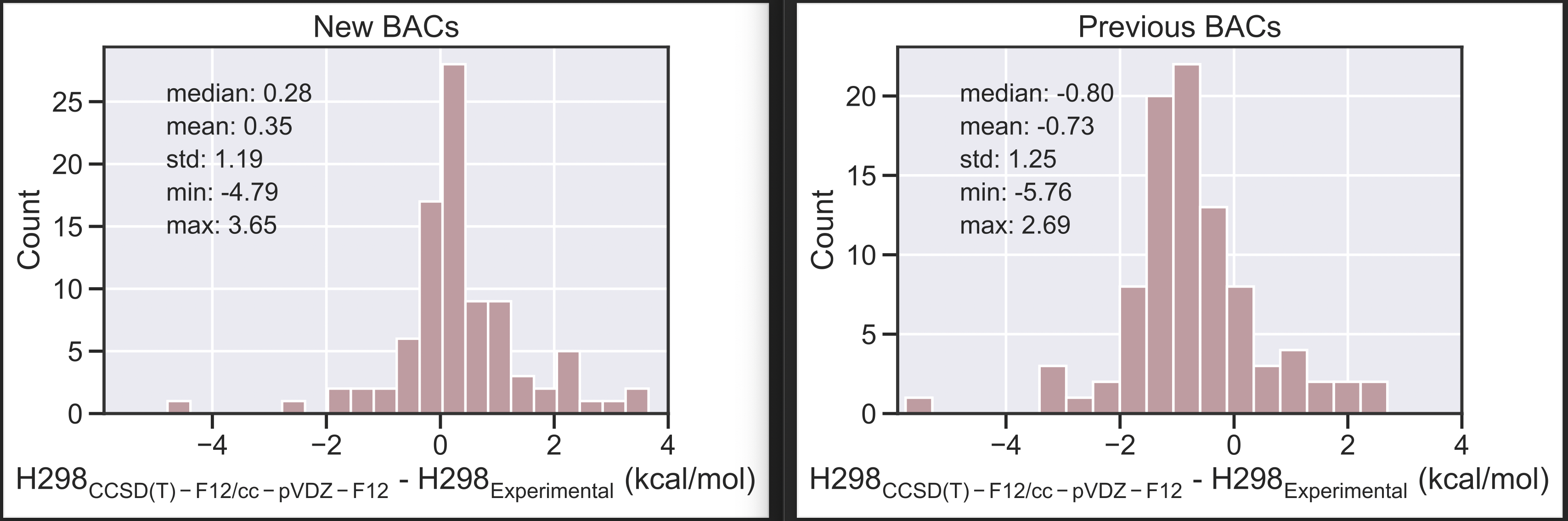
This PR updates the BACs for 4 different LoT. Previously, the BACs were calculated incorrectly due to an error with the frequency scale factor. Here, the enthalpies used for fitting BACs are calculated correctly, which slightly changes the BAC values. The numerical differences are small, but should be corrected nonetheless since they impact the estimate of corrected enthalpies.
The updated BACs improve our enthalpy estimations. For example, we can examine the error of our calculated enthalpies (composite LoT + AEC + BAC) vs. experimental values in the Pedley set, which have experimental uncertainty of less than 1 kcal/mol. As a relatively easy comparison, we can look at the error of species from the Pedley set that are also in our RMG-database reference set used for fitting the BACs. The new MAE value is 0.482 kcal/mol, which is an improvement over the previous value of 0.798 kcal/mol with the previous BACs. For context, the training MAE across all species in our reference set as 0.52 kcal/mol so these low values are expected.
As a more challenging comparison, we can also examine the error for species that were not in our reference set. This testing error represents a more realistic error that one might expect to see when using these BACs on new species. As seen in the plots below, the new BACs (left) are more centered around 0 in comparison to the old BACs (right). Consequently, the new MAE is 0.837 kcal/mol, which is an improvement over the previous value of 1.160 kcal/mol with the previous BACs.