-
Notifications
You must be signed in to change notification settings - Fork 168
Closed
Description
I am trying to run PARCELS with the output of the HYCOM hydrodynamic model.
I am able to initialise the fieldset with:
fname = 'uv3z.nc4'
filenames = {'U': fname, 'V': fname}
variables = {'U': 'water_u', 'V': 'water_v'}
dimensions = {'lat': 'lat', 'lon': 'lon', 'time': 'time','depth':'depth'} # In the GlobCurrent data the dimensions are also called 'lon', 'lat' and 'time'
fieldset = FieldSet.from_netcdf(filenames, variables, dimensions,vmin=-2000)
and define the initial locations of my particles with:
#Define the particles type and initial conditions in a ParticleSet object
sitelocs = pandas.read_csv('sites.csv')
lat = sitelocs.lat.tolist()
lon =sitelocs.lon.tolist()
sites = len(lat)
n = 5 #number of replicate releases from each site
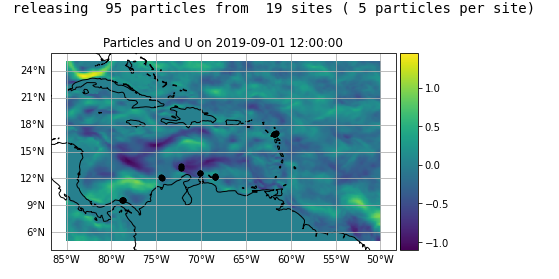
print('releasing ',n*sites,'particles from ',sites,'sites (',n,'particles per site)')
lat = list(itertools.chain.from_iterable(itertools.repeat(x, n) for x in lat))
lon = list(itertools.chain.from_iterable(itertools.repeat(x, n) for x in lon))
pset = ParticleSet(fieldset=fieldset,
pclass=JITParticle,
lon=lon, # release longitude
lat=lat) # release latitude
pset.show(field=fieldset.U)
However, when I try to execute the kernel I fall at the first hurdle:
#Execute an advection kernel on the given fieldset
output_file = pset.ParticleFile(name="reverse5.nc", outputdt=timedelta(hours=1))
pset.execute(AdvectionRK4,
runtime=timedelta(days=1),
dt=-timedelta(minutes=5),
output_file=output_file)
INFO: Compiled JITParticleAdvectionRK4 ==> /tmp/parcels-15572/75f11f043bd3a0fa57b03bbe68452681_0.so
---------------------------------------------------------------------------
OutOfBoundsError Traceback (most recent call last)
<ipython-input-4-d68316a1e96a> in <module>()
9 runtime=timedelta(days=1), # the total length of the run
10 dt=-timedelta(minutes=5), # the timestep of the kernel (negative timestep for backwards run)
---> 11 output_file=output_file)
12
~/envs/parcels/lib/python3.6/site-packages/parcels/particleset.py in execute(self, pyfunc, endtime, runtime, dt, moviedt, recovery, output_file, movie_background_field, verbose_progress, postIterationCallbacks, callbackdt)
767 time = max(next_prelease, next_input, next_output, next_movie, next_callback, endtime)
768 self.kernel.execute(self, endtime=time, dt=dt, recovery=recovery, output_file=output_file,
--> 769 execute_once=execute_once)
770 if abs(time-next_prelease) < tol:
771 pset_new = ParticleSet(fieldset=self.fieldset, time=time, lon=self.repeatlon,
~/envs/parcels/lib/python3.6/site-packages/parcels/kernel.py in execute(self, pset, endtime, dt, recovery, output_file, execute_once)
424 recovery_kernel = recovery_map[particles.state]
425 particles.set_state(StateCode.Success)
--> 426 recovery_kernel(particles, self.fieldset, particles.time)
427 if particles.state == StateCode.Success:
428 particles.set_state(StateCode.Evaluate)
~/envs/parcels/lib/python3.6/site-packages/parcels/tools/statuscodes.py in recovery_kernel_out_of_bounds(particle, fieldset, time)
184 # TODO: JIT does not yet provide the context that created
185 # the exception. We need to pass that info back from C.
--> 186 raise OutOfBoundsError(particle, fieldset)
187 else:
188 error = particle.exception
OutOfBoundsError: 0
Particle P[0](lon=-61.644100, lat=17.032516, depth=0.000000, time=10443600.000000)
Time: 2019-12-31T09:00:00.000000000, timestep dt: -300.000000
Out-of-bounds sampling by particle at (-61.644100, 17.032516, 0.000000)
Its not too clear to me why this is happening as my plot seems to suggest that my particles are well within the bounds of my model. Any insight would be greatly appreciated.
Metadata
Metadata
Assignees
Labels
No labels