Simulations and validations for simple PCA neurophysiological data alignment cleaning strategy.
This repository contains a simple tool for "cleaning" misaligned neurophysiological time-series which are meant to be event-locked to a fixed behavioral event of interest. For example, a behavior of interest may be captured using a video camera with a framerate of 30 Hz, while the relevant timescale for spiking processes of interest is orders of magnitude faster (300 - 3000 Hz). The jitter introduced by misalignment even within a single frame (by comparison to the spike timescale) is a noise source for the physiological data of interest. Fortunately, we can correct for this noise source to remove jitter from recovered single-trial spike rate estimates by implementing a simple PCA noise rejection step that aims to reconstruct the correctly-aligned data under following assumption: we can "discard" part of the data in a principled way, such that the reconstruction error contains the unwanted misalignment noise.
In order for this to be a useful utility for neurophysiological behavior analysis in preclinical studies, we would like to achieve the following:
Identify a "robust" surface manifold that relates the optimal retained data variance based on a combination of known or estimable parameters such as:
- Number of channels
- Number of trials
- Expected jitter standard deviation
- Expected non-neural noise variance
- Total number of "generative" factors present for spike rates
There are pretty much just two steps to follow:
- If not already cloned to the same folder containing this repository, add the following repository at that location (or modify
default.Repos()local path to reflect its location; note that the local name on my PC is just'Projects/Utilities', so if you clone using the default repo name ('Matlab_Utilities') make sure to reflect that): https://github.com/m053m716/Matlab_Utilities- This should be the only major dependency the repository has, and it's not really that important other than for using the
savefunctions inmain.mlx.
- This should be the only major dependency the repository has, and it's not really that important other than for using the
- Literally, just click through
main.mlxin a Matlab release that supports the MatlabLive Editor(likely,Matlab R2016aand beyond).
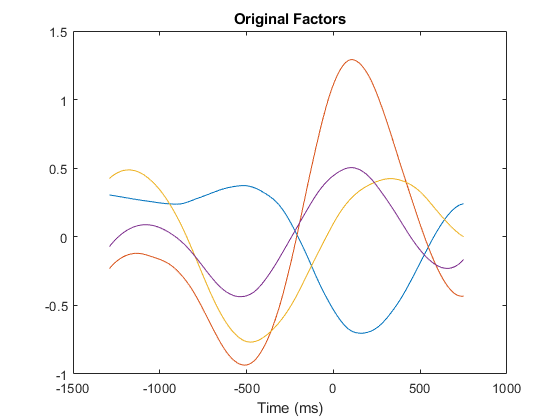
The factors used for simulation of spike trains are derived from a reach-to-grasp experiment recorded from a rat reaching to retrieve a pellet, in alignment to the first video frame on which the rat closed its paw.
The original factors are scaled as coefficients between -1 and 1.
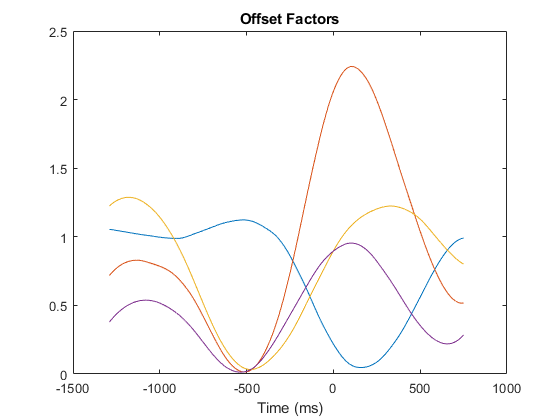
Each factor offset is just to make them non-negative for all points, so that they can be scaled to physiologically plausible spike rate parameters.
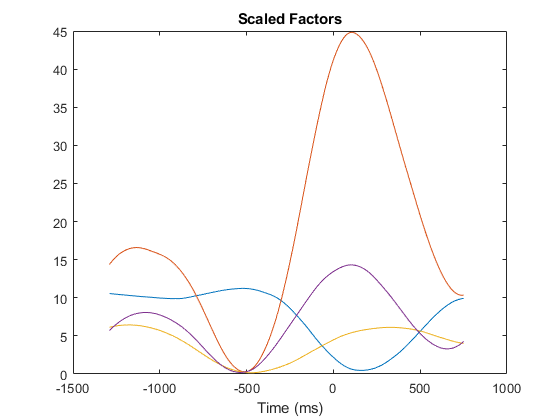
After the offset, factors are rescaled so that their units change to spikes/second. All subsequent simulations are weighted combinations of these four factors.
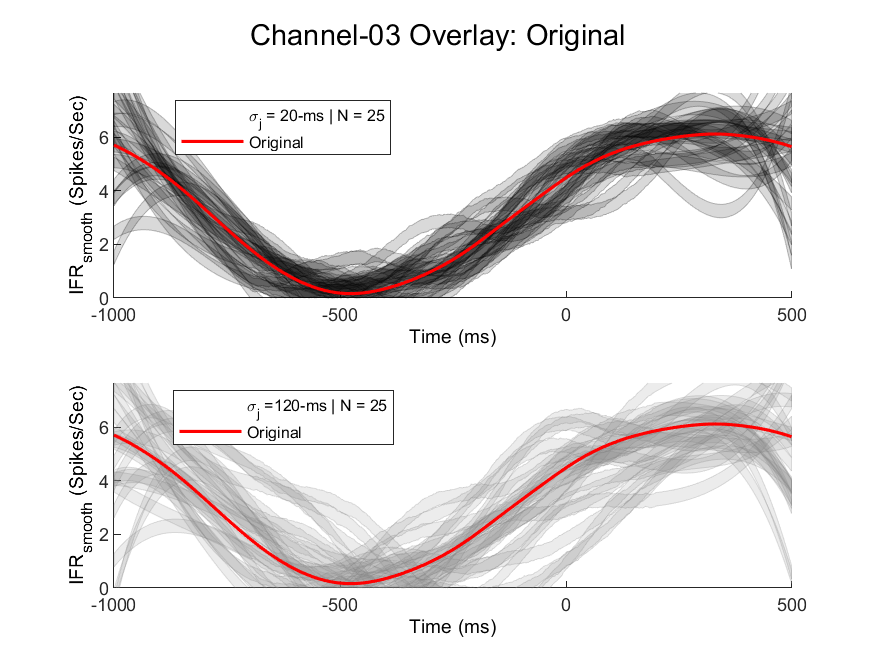
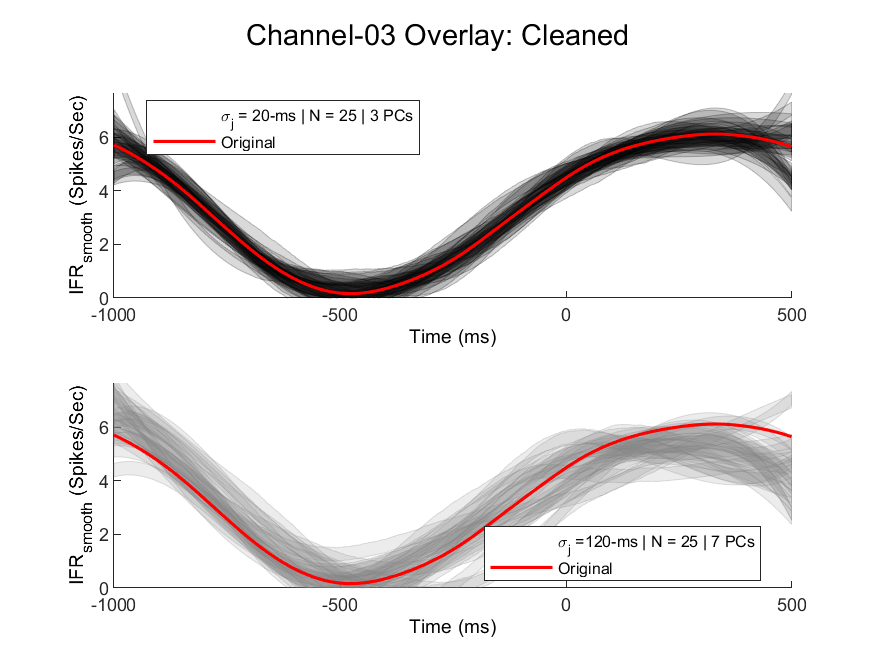
Here is an example of fairly noisy data with jitter on the order of 20-ms (low-noise case) up to 120-ms (high-noise case) for a realistic number of trials (N = 25). Each semi-opaque trajectory represents an individual trial for that recording "channel," which is some weighted combination of the scaled factors Note that particularly in rehabilitation experiments, it is difficult to get a high number of trials for a variety of reasons, but particularly if the animal belongs to an injury group they may not be very motivated to perform hundreds of behavioral trials.
Using the same simulated data, we retain the top-3 (top: low-noise) or top-7 (bottom: high-noise) principal components and reconstruct the simulated spike rates. The number of principal components is set using a threshold based on the total percent of the variance explained by the retained principal components. Part of the goal of this small project is to identify a "robust" surface manifold based on parameters such as the number of channels, number of trials, noise parameters, and total number of "generative" factors present for spike rates.