AtlasPatch: An Efficient and Scalable Tool for Whole Slide Image Preprocessing in Computational Pathology
Project Page | Paper | Hugging Face | GitHub
- Installation
- Usage Guide
- Supported Formats
- Using Extracted Data
- Available Feature Extractors
- Bring Your Own Encoder
- SLURM job scripts
- Frequently Asked Questions (FAQ)
- Feedback
- Citation
- License
- Future Updates
# Install AtlasPatch
pip install atlas-patch
# Install SAM2 (required for tissue segmentation)
pip install git+https://github.com/facebookresearch/sam2.gitNote: AtlasPatch requires the OpenSlide system library for WSI processing. See OpenSlide Prerequisites below.
Before installing AtlasPatch, you need the OpenSlide system library:
-
Using Conda (Recommended):
conda install -c conda-forge openslide
-
Ubuntu/Debian:
sudo apt-get install openslide-tools
-
macOS:
brew install openslide
-
Other systems: Visit OpenSlide Documentation
Some feature extractors require additional dependencies that must be installed separately:
# For CONCH encoder (conch_v1, conch_v15)
pip install git+https://github.com/Mahmoodlab/CONCH.git
# For MUSK encoder
pip install git+https://github.com/lilab-stanford/MUSK.gitThese are only needed if you plan to use those specific encoders.
Using Conda Environment
# Create and activate environment
conda create -n atlas_patch python=3.10
conda activate atlas_patch
# Install OpenSlide
conda install -c conda-forge openslide
# Install AtlasPatch and SAM2
pip install atlas-patch
pip install git+https://github.com/facebookresearch/sam2.gitUsing uv (faster installs)
# Install uv (see https://docs.astral.sh/uv/getting-started/)
curl -LsSf https://astral.sh/uv/install.sh | sh
# Create and activate environment
uv venv
source .venv/bin/activate # On Windows: .venv\Scripts\activate
# Install AtlasPatch and SAM2
uv pip install atlas-patch
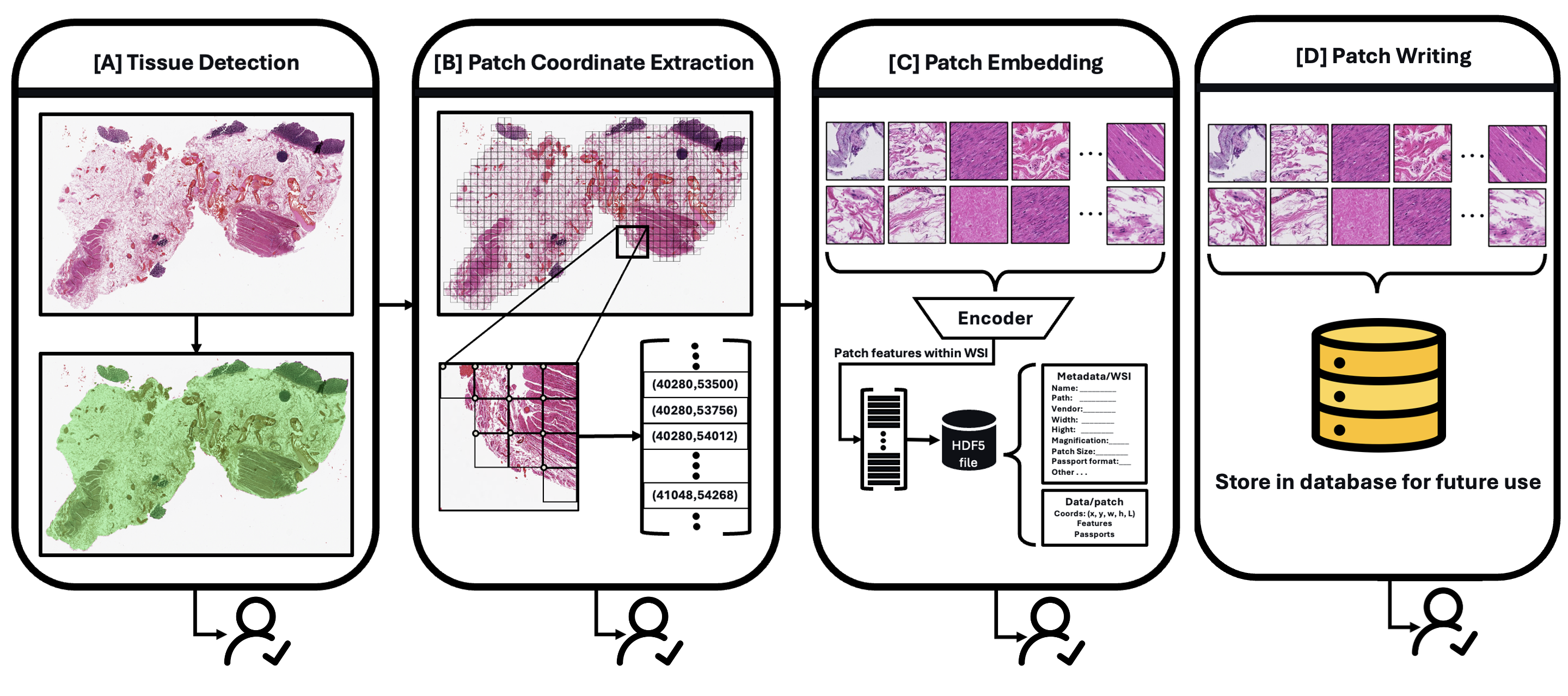
uv pip install git+https://github.com/facebookresearch/sam2.gitAtlasPatch provides a flexible pipeline with 4 checkpoints that you can use independently or combine based on your needs.
Quick overview of the checkpoint commands:
detect-tissue: runs SAM2 segmentation and writes mask overlays under<output>/visualization/.segment-and-get-coords: runs segmentation + patch coordinate extraction into<output>/patches/<stem>.h5.process: full pipeline (segmentation + coords + feature embeddings) in the same H5.segment-and-get-coords --save-images: same assegment-and-get-coords, plus patch PNGs under<output>/images/<stem>/.
Detect and visualize tissue regions in your WSI using SAM2 segmentation.
atlaspatch detect-tissue /path/to/slide.svs \
--output ./output \
--device cudaDetect tissue and extract patch coordinates without feature embedding.
atlaspatch segment-and-get-coords /path/to/slide.svs \
--output ./output \
--patch-size 256 \
--target-mag 20 \
--device cudaRun the full pipeline: Tissue detection, coordinate extraction, and feature embedding.
atlaspatch process /path/to/slide.svs \
--output ./output \
--patch-size 256 \
--target-mag 20 \
--feature-extractors resnet50 \
--device cudaFull pipeline with optional patch image export for visualization or downstream tasks.
atlaspatch segment-and-get-coords /path/to/slide.svs \
--output ./output \
--patch-size 256 \
--target-mag 20 \
--device cuda \
--save-imagesPass a directory instead of a single file to process multiple WSIs; outputs land in <output>/patches/<stem>.h5 based on the path you provide to --output.
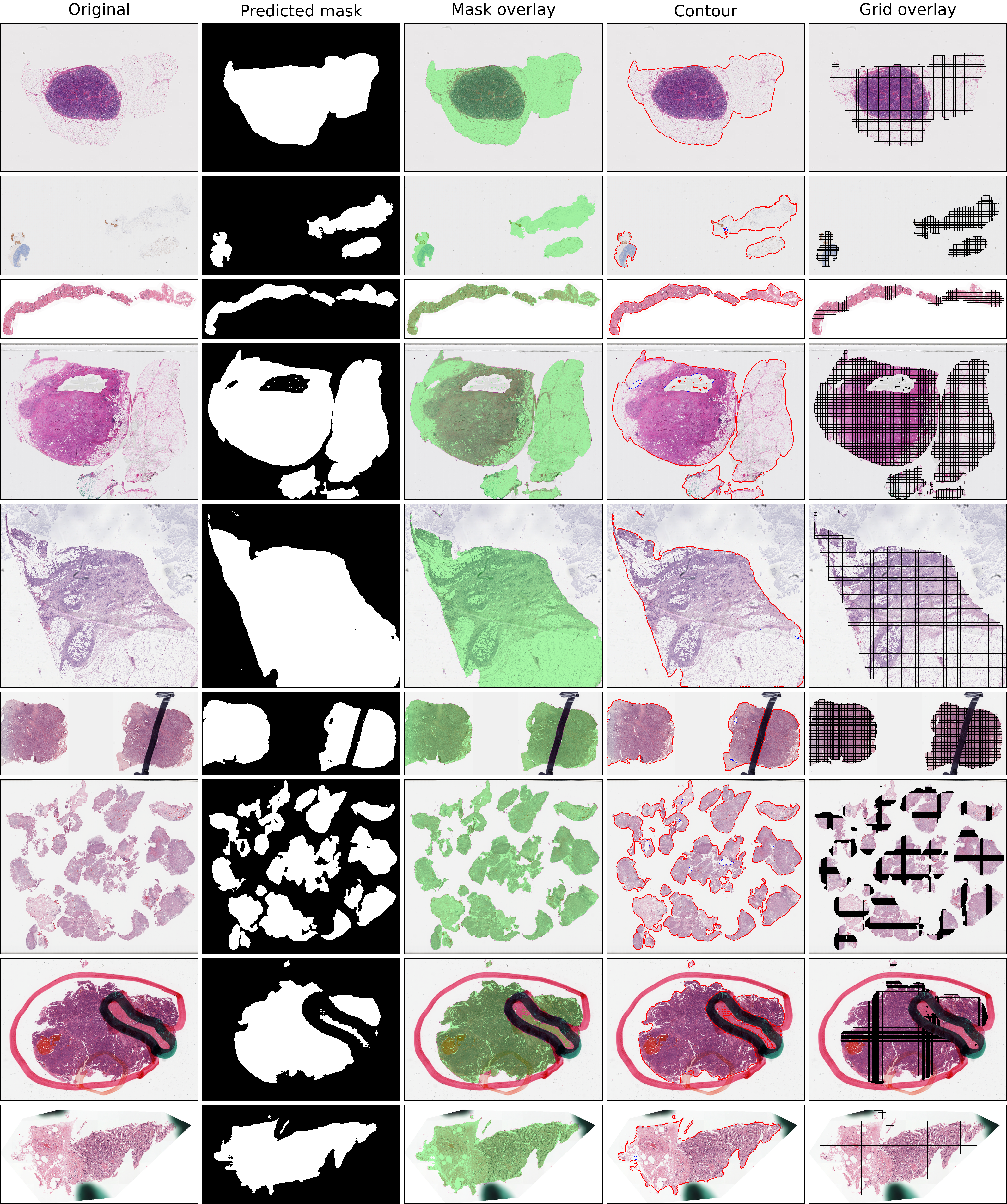
Below are some examples for the output masks and overlays (original image, predicted mask, overlay, contours, grid).
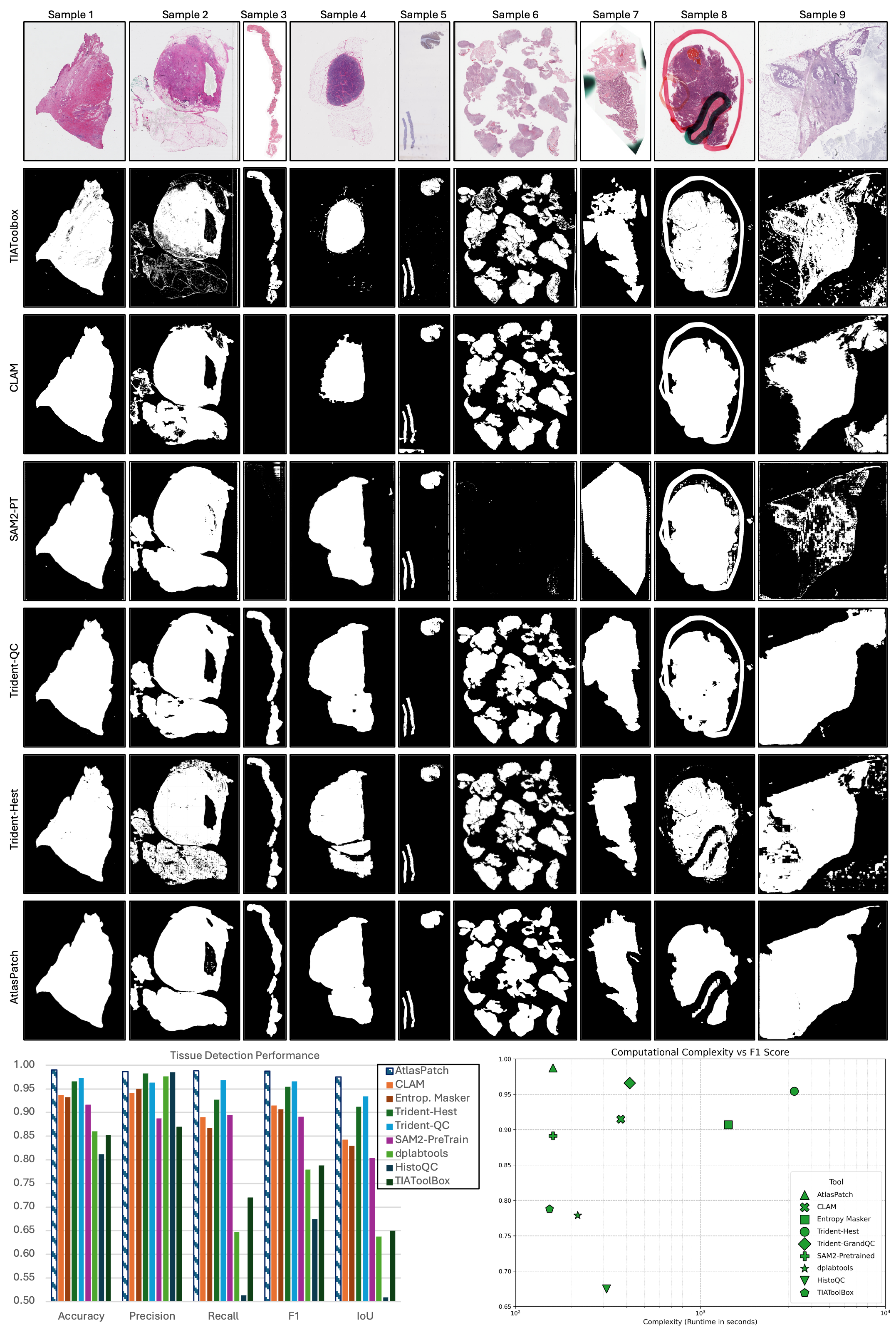
Quantitative and qualitative analysis of AtlasPatch tissue detection against existing slide-preprocessing tools.
Representative WSI thumbnails are shown from diverse tissue features and artifact conditions, with tissue masks predicted by thresholding methods (TIAToolbox, CLAM) and deep learning methods (pretrained "non-finetuned" SAM2 model, Trident-QC, Trident-Hest and AtlasPatch), highlighting differences in boundary fidelity, artifact suppression and handling of fragmented tissue (more tools are shown in the appendix). Tissue detection performance is also shown on the held-out test set for AtlasPatch and baseline pipelines, highlighting that AtlasPatch matches or exceeds their segmentation quality. The segmentation complexity–performance trade-off, plotting F1-score against segmentation runtime (on a random set of 100 WSIs), shows AtlasPatch achieves high performance with substantially lower wall-clock time than tile-wise detectors and heuristic pipelines, underscoring its suitability for large-scale WSI preprocessing.
The process command is the primary entry point for most workflows. It runs the full pipeline: tissue segmentation, patch coordinate extraction, and feature embedding. You can process a single slide or an entire directory of WSIs in one command.
atlaspatch process <WSI_PATH> --output <DIR> --patch-size <INT> --target-mag <INT> --feature-extractors <NAMES> [OPTIONS]| Argument | Description |
|---|---|
WSI_PATH |
Path to a single slide file or a directory containing slides. When a directory is provided, all supported formats are processed. |
--output, -o |
Root directory for results. Outputs are organized as <output>/patches/<stem>.h5 for coordinates and features, and <output>/visualization/ for overlays. |
--patch-size |
Final patch size in pixels at the target magnification (e.g., 256 for 256×256 patches). |
--target-mag |
Magnification level to extract patches at. Common values: 5, 10, 20, 40. The pipeline reads from the closest available pyramid level and resizes if needed. |
--feature-extractors |
Comma or space-separated list of encoder names from Available Feature Extractors. Multiple encoders can be specified to extract several feature sets in one pass (e.g., resnet50,uni_v2). |
| Argument | Default | Description |
|---|---|---|
--step-size |
Same as --patch-size |
Stride between patches. Omit for non-overlapping grids. Use smaller values (e.g., 128 with --patch-size 256) to create 50% overlap. |
| Argument | Default | Description |
|---|---|---|
--device |
cuda |
Device for SAM2 tissue segmentation. Options: cuda, cuda:0, cuda:1, or cpu. |
--seg-batch-size |
1 |
Batch size for SAM2 thumbnail segmentation. Increase for faster processing if GPU memory allows. |
--patch-workers |
CPU count | Number of threads for patch extraction and H5 writes. |
--max-open-slides |
200 |
Maximum number of WSI files open simultaneously. Lower this if you hit file descriptor limits. |
| Argument | Default | Description |
|---|---|---|
--feature-device |
Same as --device |
Device for feature extraction. Set separately to use a different GPU than segmentation. |
--feature-batch-size |
32 |
Batch size for the feature extractor forward pass. Increase for faster throughput; decrease if running out of GPU memory. |
--feature-num-workers |
4 |
Number of DataLoader workers for loading patches during feature extraction. |
--feature-precision |
float16 |
Precision for feature extraction. Options: float32, float16, bfloat16. Lower precision reduces memory and can improve throughput on compatible GPUs. |
| Argument | Default | Description |
|---|---|---|
--fast-mode |
Enabled | Skips per-patch black/white content filtering for faster processing. Use --no-fast-mode to enable filtering. |
--tissue-thresh |
0.0 |
Minimum tissue area fraction to keep a region. Filters out tiny tissue fragments. |
--white-thresh |
15 |
Saturation threshold for white patch filtering (only with --no-fast-mode). Lower values are stricter. |
--black-thresh |
50 |
RGB threshold for black/dark patch filtering (only with --no-fast-mode). Higher values are stricter. |
| Argument | Default | Description |
|---|---|---|
--visualize-grids |
Off | Render patch grid overlay on slide thumbnails. |
--visualize-mask |
Off | Render tissue segmentation mask overlay. |
--visualize-contours |
Off | Render tissue contour overlay. |
All visualization outputs are saved under <output>/visualization/.
| Argument | Default | Description |
|---|---|---|
--save-images |
Off | Export each patch as a PNG file under <output>/images/<stem>/. |
--recursive |
Off | Walk subdirectories when WSI_PATH is a directory. |
--mpp-csv |
None | Path to a CSV file with wsi,mpp columns to override microns-per-pixel when slide metadata is missing or incorrect. |
--skip-existing |
Off | Skip slides that already have an output H5 file. |
--force |
Off | Overwrite existing output files. |
--verbose, -v |
Off | Enable debug logging and disable the progress bar. |
--write-batch |
8192 |
Number of coordinate rows to buffer before flushing to H5. Tune for RAM vs. I/O trade-off. |
AtlasPatch uses OpenSlide for WSIs and Pillow for standard images:
- WSIs:
.svs,.tif,.tiff,.ndpi,.vms,.vmu,.scn,.mrxs,.bif,.dcm - Images:
.png,.jpg,.jpeg,.bmp,.webp,.gif
atlaspatch process writes one HDF5 per slide under <output>/patches/<stem>.h5 containing coordinates and feature matrices. Coordinates and features share row order.
- Dataset:
coords(int32, shape(N, 5)) with columns(x, y, read_w, read_h, level). xandyare level-0 pixel coordinates.read_w,read_h, andleveldescribe how the patch was read from the WSI.- The level-0 footprint of each patch is stored as the
patch_size_level0file attribute; some slide encoders use it for positional encoding (e.g., ALiBi in TITAN).
Example:
import h5py
import numpy as np
import openslide
from PIL import Image
h5_path = "output/patches/sample.h5"
wsi_path = "/path/to/slide.svs"
with h5py.File(h5_path, "r") as f:
coords = f["coords"][...] # (N, 5) int32: [x, y, read_w, read_h, level]
patch_size = int(f.attrs["patch_size"])
with openslide.OpenSlide(wsi_path) as wsi:
for x, y, read_w, read_h, level in coords:
img = wsi.read_region(
(int(x), int(y)),
int(level),
(int(read_w), int(read_h)),
).convert("RGB")
if img.size != (patch_size, patch_size):
# Some slides don't have a pyramid level that matches target magnification exactly, so they have to be resized.
img = img.resize((patch_size, patch_size), resample=Image.BILINEAR)
patch = np.array(img) # (H, W, 3) uint8- Group:
features/inside the same HDF5. - Each extractor is stored as
features/<name>(float32, shape(N, D)), aligned row-for-row withcoords. - List available feature sets with
list(f['features'].keys()).
import h5py
with h5py.File("output/patches/sample.h5", "r") as f:
feat_names = list(f["features"].keys())
resnet50_feats = f["features/resnet50"][...] # (N, 2048) float32| Name | Output Dim |
|---|---|
resnet18 |
512 |
resnet34 |
512 |
resnet50 |
2048 |
resnet101 |
2048 |
resnet152 |
2048 |
convnext_tiny |
768 |
convnext_small |
768 |
convnext_base |
1024 |
convnext_large |
1536 |
vit_b_16 |
768 |
vit_b_32 |
768 |
vit_l_16 |
1024 |
vit_l_32 |
1024 |
vit_h_14 |
1280 |
dinov2_small (DINOv2: Learning Robust Visual Features without Supervision) |
384 |
dinov2_base (DINOv2: Learning Robust Visual Features without Supervision) |
768 |
dinov2_large (DINOv2: Learning Robust Visual Features without Supervision) |
1024 |
dinov2_giant (DINOv2: Learning Robust Visual Features without Supervision) |
1536 |
dinov3_vits16 (DINOv3) |
384 |
dinov3_vits16_plus (DINOv3) |
384 |
dinov3_vitb16 (DINOv3) |
768 |
dinov3_vitl16 (DINOv3) |
1024 |
dinov3_vitl16_sat (DINOv3) |
1024 |
dinov3_vith16_plus (DINOv3) |
1280 |
dinov3_vit7b16 (DINOv3) |
4096 |
dinov3_vit7b16_sat (DINOv3) |
4096 |
Note: Some encoders (e.g.,
uni_v1, etc.) require access approval from Hugging Face. To use these models:
- Request access on the respective Hugging Face model page
- Once approved, set your Hugging Face token as an environment variable:
export HF_TOKEN=your_huggingface_token- Then you can use the encoder in your commands
| Name | Output Dim |
|---|---|
clip_rn50 (Learning Transferable Visual Models From Natural Language Supervision) |
1024 |
clip_rn101 (Learning Transferable Visual Models From Natural Language Supervision) |
512 |
clip_rn50x4 (Learning Transferable Visual Models From Natural Language Supervision) |
640 |
clip_rn50x16 (Learning Transferable Visual Models From Natural Language Supervision) |
768 |
clip_rn50x64 (Learning Transferable Visual Models From Natural Language Supervision) |
1024 |
clip_vit_b_32 (Learning Transferable Visual Models From Natural Language Supervision) |
512 |
clip_vit_b_16 (Learning Transferable Visual Models From Natural Language Supervision) |
512 |
clip_vit_l_14 (Learning Transferable Visual Models From Natural Language Supervision) |
768 |
clip_vit_l_14_336 (Learning Transferable Visual Models From Natural Language Supervision) |
768 |
Add a custom encoder without touching AtlasPatch by writing a small plugin and pointing the CLI at it with --feature-plugin /path/to/plugin.py. The plugin must expose a register_feature_extractors(registry, device, dtype, num_workers) function; inside that hook call register_custom_encoder with a loader that knows how to load the model and run a forward pass.
import torch
from torchvision import transforms
from atlas_patch.models.patch.custom import CustomEncoderComponents, register_custom_encoder
def build_my_encoder(device: torch.device, dtype: torch.dtype) -> CustomEncoderComponents:
"""
Build the components used by AtlasPatch to embed patches with a custom model.
Returns:
CustomEncoderComponents describing the model, preprocess transform, and forward pass.
"""
model = ... # your torch.nn.Module
model = model.to(device=device, dtype=dtype).eval()
preprocess = transforms.Compose([transforms.Resize(224), transforms.ToTensor()])
def forward(batch: torch.Tensor) -> torch.Tensor:
return model(batch) # must return [batch, embedding_dim]
return CustomEncoderComponents(model=model, preprocess=preprocess, forward_fn=forward)
def register_feature_extractors(registry, device, dtype, num_workers):
register_custom_encoder(
registry=registry,
name="my_encoder",
embedding_dim=512,
loader=build_my_encoder,
device=device,
dtype=dtype,
num_workers=num_workers,
)Run AtlasPatch with --feature-plugin /path/to/plugin.py --feature-extractors my_encoder to benchmark your encoder alongside the built-ins, multiple plugins and extractors can be added at once. Outputs keep the same HDF5 layout—your custom embeddings live under features/my_encoder (row-aligned with coords) next to other extractors.
We prepared ready-to-run SLURM templates under jobs/:
- Patch extraction (SAM2 + H5/PNG):
jobs/atlaspatch_patch.slurm.sh. Edits to make:- Set
WSI_ROOT,OUTPUT_ROOT,PATCH_SIZE,TARGET_MAG,SEG_BATCH. - Ensure
--cpus-per-taskmatches the CPU you want; the script passes--patch-workers ${SLURM_CPUS_PER_TASK}and caps--max-open-slidesat 200. --fast-modeis on by default; append--no-fast-modeto enable content filtering.- Submit with
sbatch jobs/atlaspatch_patch.slurm.sh.
- Set
- Feature embedding (adds features into existing H5 files):
jobs/atlaspatch_features.slurm.sh. Edits to make:- Set
WSI_ROOT,OUTPUT_ROOT,PATCH_SIZE, andTARGET_MAG. - Configure
FEATURES(comma/space list, multiple extractors are supported),FEATURE_DEVICE,FEATURE_BATCH,FEATURE_WORKERS, andFEATURE_PRECISION. - This script is intended for feature extraction; use the patch script when you need segmentation + coordinates, and run the feature script to embed one or more models into those H5 files.
- Submit with
sbatch jobs/atlaspatch_features.slurm.sh.
- Set
- Running multiple jobs: you can submit several jobs in a loop (e.g., 50 job using
for i in {1..50}; do sbatch jobs/atlaspatch_features.slurm.sh; done). AtlasPatch uses per-slide lock files to avoid overlapping work on the same slide.
I'm facing an out of memory (OOM) error
This usually happens when too many WSI files are open simultaneously. Try reducing the --max-open-slides parameter:
atlaspatch process /path/to/slides --output ./output --max-open-slides 50The default is 200. Lower this value if you're processing many large slides or have limited system memory.
I'm getting a CUDA out of memory error
Try one or more of the following:
-
Reduce feature extraction batch size:
--feature-batch-size 16 # Default is 32 -
Reduce segmentation batch size:
--seg-batch-size 1 # Default is 1 -
Use lower precision:
--feature-precision float16 # or bfloat16 -
Use a smaller patch size:
--patch-size 224 # Instead of 256
OpenSlide library not found
AtlasPatch requires the OpenSlide system library. Install it based on your system:
- Conda:
conda install -c conda-forge openslide - Ubuntu/Debian:
sudo apt-get install openslide-tools - macOS:
brew install openslide
See OpenSlide Prerequisites for more details.
Access denied for gated models (UNI, Virchow, etc.)
Some encoders require Hugging Face access approval:
- Request access on the model's Hugging Face page (e.g., UNI)
- Once approved, set your token:
export HF_TOKEN=your_huggingface_token - Run AtlasPatch again
Missing microns-per-pixel (MPP) metadata
Some slides lack MPP metadata. You can provide it via a CSV file:
atlaspatch process /path/to/slides --output ./output --mpp-csv /path/to/mpp.csvThe CSV should have columns wsi (filename) and mpp (microns per pixel value).
Processing is slow
Try these optimizations:
-
Enable fast mode (skips content filtering, enabled by default):
--fast-mode
-
Increase parallel workers:
--patch-workers 16 # Match your CPU cores --feature-num-workers 8 -
Increase batch sizes (if GPU memory allows):
--feature-batch-size 64 --seg-batch-size 4
-
Use multiple GPUs by running separate jobs on different GPU devices.
My file format is not supported
AtlasPatch supports most common formats via OpenSlide and Pillow:
- WSIs:
.svs,.tif,.tiff,.ndpi,.vms,.vmu,.scn,.mrxs,.bif,.dcm - Images:
.png,.jpg,.jpeg,.bmp,.webp,.gif
If your format isn't supported, consider converting it to a supported format or open an issue.
How do I skip already processed slides?
Use the --skip-existing flag to skip slides that already have an output H5 file:
atlaspatch process /path/to/slides --output ./output --skip-existingHave a question not covered here? Feel free to open an issue and ask!
- Report problems via the bug report template so we can reproduce and fix them quickly.
- Suggest enhancements through the feature request template with your use case and proposal.
- When opening a PR, fill out the pull request template and run the listed checks (lint, format, type-check, tests).
If you use AtlasPatch in your research, please cite our paper:
@article{atlaspatch2025,
title = {AtlasPatch: An Efficient and Scalable Tool for Whole Slide Image Preprocessing in Computational Pathology},
author = {Alagha, Ahmed and Leclerc, Christopher and Kotp, Yousef and Abdelwahed, Omar and Moras, Calvin and Rentopoulos, Peter and Rostami, Rose and Nguyen, Bich Ngoc and Baig, Jumanah and Khellaf, Abdelhakim and Trinh, Vincent Quoc-Huy and Mizouni, Rabeb and Otrok, Hadi and Bentahar, Jamal and Hosseini, Mahdi S.},
journal = {arXiv},
year = {2025},
url = {TODO: coming soon}
}AtlasPatch is released under CC-BY-NC-SA-4.0, which strictly disallows commercial use of the model weights or any derivative works. Commercialization includes selling the model, offering it as a paid service, using it inside commercial products, or distributing modified versions for commercial gain. Non-commercial research, experimentation, educational use, and use by academic or non-profit organizations is permitted under the license terms. If you need commercial rights, please contact the authors to obtain a separate commercial license. See the LICENSE file in this repository for full terms. For the complete license text and detailed terms, see the LICENSE file in this repository.
- We plan to add slide-level encoders (open for extension): TITAN, PRISM, GigaPath, Madeleine.