Harnessing multiple LLMs for Information Retrieval A case study on Deep Learning methodologies in Biodiversity publications
source: https://arxiv.org/html/2411.09269v1 by Vamsi Krishna Kommineni, Birgitta König-Ries, Sheeba Samuel
- Abstract
- Introduction
- Related Work
- Methods
- Results
- Discussion
- Conclusions
- Acknowledgments
- Data availability statement
Deep Learning (DL) Techniques in Scientific Studies
- Increasingly applied: DL techniques used across various domains to address complex research questions
- Methodological details hidden: Details of DL models are often unstructured and difficult to access/comprehend
Addressing the Issue
- Used five different open-source Large Language Models (LLMs): Llama-3 70B, Llama-3.1 70B, Mixtral-8x22B-Instruct-v0.1, Mixtral 8x7B, and Gemma 2 9B
- Implemented Retrieval-Augmented Generation (RAG) approach to automatically extract DL methodological details from scientific publications
Voting Classifier for DL Methodological Information
- Built a voting classifier from the outputs of the five LLMs
- Designed to accurately report DL methodological information
Testing and Validation
- Used two datasets of DL-related biodiversity publications:
- Curated set of 100 publications from previous work
- Additional set of 364 publications from the Ecological Informatics journal
- Results demonstrated that the multi-LLM, RAG-assisted pipeline enhances retrieval of DL methodological information:
- Achieved an accuracy of 69.5% (417 out of 600 comparisons) based on textual content alone
- Assessed against human annotators with access to codes, figures, tables, etc.
Limitations and Future Work
- Methodology not limited to biodiversity; can be applied across other scientific domains
- Enhances knowledge transfer and ensures better reproducibility
Deep Learning (DL)
- Cornerstone in numerous fields, revolutionizing complex data analysis and interpretation
- Applications: healthcare, finance, autonomous systems, natural language processing
- Increasing recognition of critical shortcoming: limited availability of detailed methodological information in scientific literature [^41]
Challenges with DL Pipelines
- Need for transparent reporting to support replication and research advancement [^14]
- Detailed documentation essential for reproducible DL pipelines [^35]
- Includes logging data collection methods, preprocessing steps, model architecture configurations, hyperparameters, and training details
- Performance metrics and test datasets, software libraries, hardware, frameworks, and versions also important
Importance of Transparency in DL Applications
- Influences real-world decisions (healthcare, finance)
- Absence of methodological transparency can compromise trust in DL models [^22]
Automated Extraction of DL Methodological Information
- Traditional manual retrieval methods inconsistent [^30]
- Proposed approach: utilize Large Language Models (LLMs) for automated extraction and processing of DL methodological information from scientific publications
LLM Models Used in Study
- Llama-3 70B11
- Llama-3.1 70B22
- Mixtral-8x22B-Instruct-v0.133
- Mixtral 8x7B44
- Gemma 2 9B55
Methodology Components
- Identifying relevant research publications
- Automatically extracting information through Retrieval-Augmented Generation (RAG) approach
- Converting extracted textual responses into categorical values for evaluation
Application to Biodiversity Publications
- Growing popularity of DL methods in biodiversity research
- Enormous number of publications using DL for various applications
- Importance of transparent sharing of DL information [^17]
Voting Classifier for Reliability
- Aggregates outputs of multiple LLMs to ensure reported information is consistent and accurate
Evaluation
- Applied to two distinct datasets: 100 publications from previous work [^5], 364 publications from Ecological Informatics journal6
Contribution to Research
- Enhances transparency and reproducibility of research by identifying gaps in reporting and ensuring critical DL methodology information is accessible.
Deep Learning Integration in Scientific Research
- DL has led to significant advancements in data analysis, pattern recognition, and predictive modeling across various fields
- Importance of transparency and replicability in DL research has been emphasized due to gaps in methodological reporting
- Lack of detailed documentation of DL methodologies is a concern in multiple domains, including biodiversity research
- Inadequate documentation hampers replication and building upon findings for stakeholders like conservationists and ecologists
- Need for comprehensive and transparent methodological documentation is crucial for reproducibility
Deep Learning in Biodiversity Research
- DL has seen rapid adoption in biodiversity due to its ability to handle large-scale, complex ecological data
- Applications include species identification, habitat classification, population monitoring (e.g., [^10], [^33])
- Transparency in methods is essential for replicability and enabling further advancements
Challenges in Documenting DL Methodologies
- Rapidly evolving field with new techniques emerging, making it difficult to keep up with reporting standards
- Complexity of ecological data and interdisciplinary nature of biodiversity research complicates documentation ([^14], [^32])
- Previous calls for better documentation practices and standardization ([^20], [^35])
- Data archiving with clear metadata essential to enhance reproducibility ([^42], [^32])
- Recent efforts focus on creating reproducible workflows for studies involving complex DL techniques
Leveraging Large Language Models (LLMs) in Methodological Extraction
- LLMs, such as GPT-3 and successors, demonstrate remarkable abilities in natural language understanding and generation
- Recent studies explore combining LLMs with retrieval mechanisms to enhance information extraction (Retrieval-Augmented Generation or RAG)
- Our study utilizes a multi-LLM and RAG-based pipeline to systematically retrieve and categorize DL-related methodological details from scientific articles.
Environmental Impact of Computational Processes in DL Research
- Training LLMs and executing complex pipelines consume substantial energy, contributing to carbon emissions ([^28])
- Call for increased awareness of ecological impact and assessing environmental footprints as part of research best practices
- Our study aims to quantify the environmental footprint of the DL-powered information retrieval pipeline.
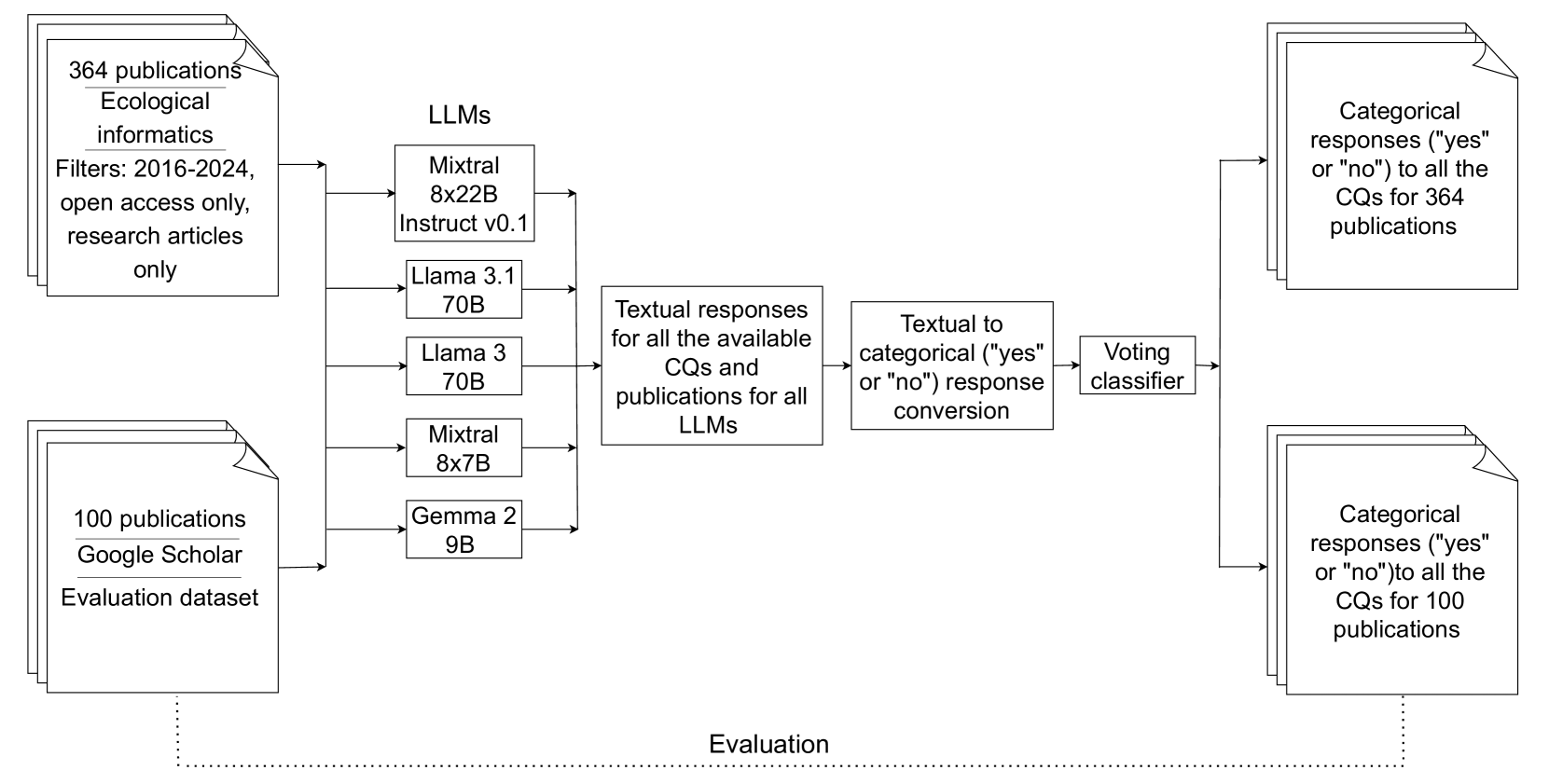
We describe a pipeline to extract and analyze biodiversity-related publication data.
Figure 1: Our workflow consists of a main process flow (solid arrows), with an evaluation phase (dotted line) for categorical responses from our previous research.
Dataset 1: Previous Research
- Indexed over 8000 results from Google Scholar using a modified version of keywords from previous research
- Narrowed down selection to 100 publications after excluding conference abstracts, theses, books, summaries, and preprints
- Manually extracted deep learning information on ten variables: Dataset, Source Code, Open source frameworks or environment, Model architecture, Software and Hardware Specification, Methods, Hyperparameters, Randomness, Averaging result, and Evaluation metrics
- These 100 publications serve as an evaluation dataset for the current study
Dataset 2: Ecological Informatics Journal
- Identified relevant deep learning keywords from AI-related session abstracts at Biodiversity Information Standards (TDWG) conferences in 2018, 2019, and 2021–2023
- Used an open-source large language model (Mixtral 8x22b Instruct-v0.1) to extract deep learning-related keywords from the abstracts
- Reduced redundancies and non-deep-learning-related terms through LLM prompting and domain expert curation, resulting in 25 optimized keywords: acoustic model, artificial intelligence, species identification, CNN model, convolutional neural network, convolutional vision transformer, deep learning, deep model, generative ai, handwritten text recognition, instance segmentation, large language model, machine learning, metric learning, natural language processing, neural network, object detection, object segmentation, optical character recognition, self-supervised learning, supervised learning, transfer learning, transformer, unsupervised learning, vision transformer
- Queried Ecological Informatics journal using the 25 refined keywords, filters: publication years from 2016 to August 1, 2024, article type as research articles, and open-access availability
- Manually exported 364 unique publications in BibTeX format after removing duplicates based on DOIs
- Retrieved full-text PDFs of the 364 unique publications through Elsevier API for further analysis.
We employed competency questions to retrieve specific deep learning methodological info from 464 biodiversity papers using multiple Large Language Models. The same 28 CQs used in our prior work were applied here.
Recent Applications of RAG Approach
- Rapidly used for information retrieval from structured and unstructured data
- Leverages large language models (LLMs) to extract information from authoritative sources, such as biodiversity publications
- Employed five LLMs: Mixtral 8x22B Instruct v0.1, Llama 3.1 70B, Llama 3 70B, Mixtral 8x7B, and Gemma 2 9B
- Temperature set to 0 for all models
- Custom GPU used for some models, API access for others
Previous Work Pipeline
- Component of a semi-automated construction of the Knowledge Graph (KG) pipeline
- Extracts, organizes, and links information from unstructured text into structured data within the KG framework
- Streamlines process of mapping complex domain knowledge for areas like biodiversity and deep learning methodologies
- Limited retrieval tokens to 1200, chunk size to 1000, and overlap to 50 chunks
- Responses should be concise and limited to fewer than 400 words
- Output in textual sentence format for each combination of CQs and biodiversity-related publications
Improvements from Previous Information Retrieval Component
- Reduced response length to enhance clarity and focus.
Preprocessing LLM Responses
- Obtained answers to Cognitive Questionnaire (CQ) for each combination of Language Model (LLM), CQ, and publication
- Removed unnecessary structured information using Python script
- Indexed relevant information following "Helpful Answer::" or "Answer::" for Mixtral 8x22B Instruct v0.1 model
- Converted LLM textual responses into categorical "yes" or "no" answers
Prompt for Converting Textual to Categorical Responses
- Determine if answer contains specific information from a research paper based on question and answer
- Provide binary response: "yes" if specific details are provided, "no" if answer is general or lacks sourced information.
Examples: Question: What methods are utilized for collecting raw data in the deep learning pipeline (e.g., surveys, sensors, public datasets)? Answer: No explicit mention of data collection methods from research paper.
Question: What data formats are used in the deep learning pipeline (e.g., image, audio, video, CSV)? Answer: Study uses audio data for input to Convolutional Neural Network (CNN) model.
Question: What are the data annotation techniques used in the deep learning pipeline (e.g., bounding box annotation, instance segmentation)? Answer: No explicit mention of specific annotation techniques in research paper, but CNNs were employed for image classification tasks and pattern recognition.
We used multiple LLMs to retrieve DL-related info and processed it into categorical values, allowing us to build a voting classifier. A hard voting methodology was employed, where each of the five instances produced "yes" or "no" outcomes for each CQ and publication combination. The classifier made decisions based on the majority vote, improving overall result quality.
We manually evaluated all key outputs generated by the LLMs, including CQ answers and categorical responses. For CQ answers, we relied on our previous work, where 30 publications were manually evaluated. We also selected 30 random publications for each LLM, annotated their question-answer pairs, and compared them using Cohen's kappa score to measure inter-annotator agreement.
Pipeline Filtering
- RAG-assisted LLM pipeline used to identify publications containing only DL-related keywords, not actual DL pipelines
- Compared LLM findings with 100 articles focused on DL methods
- Evaluated the outputs of all publications and filtered ones
Time Logs
- Computational tasks rely on physical resources, with growing environmental footprint awareness
- Recorded time taken to process requests for each document
- Preprocessed logs by removing duplicates based on unique identifiers
- Essential for calculating the environmental footprint of DL-driven information retrieval pipeline
Semantic Similarity Between LLM Outputs
- Five textual answers per combination of CQ and publication, converted to categorical forms
- Computed cosine similarity matrix between all LLM combinations
- Assessed inter-annotator agreement among categorical responses using Cohen's kappa score
Environmental Footprint
- Recorded processing times for each publication and key pipeline components (RAG answers, conversion to categorical responses)
- Used hardware configuration, total runtime, and location data to estimate environmental footprint using a website
- Pipeline consumed 177.55 kWh of energy and 60.14 kg CO2e (equivalent to 64.65 tree months) for RAG textual responses
- Consumed 50.63 kWh, 17.15 kg CO2e, and 18.7 tree months for converting textual to categorical responses
Evaluation of LLM Responses
- Comparisons per LLM: 840 (30 publications x 28 CQs)
- Total comparisons for five LLMs: 4,200
- Agreements between LLM responses and human-annotated ground truth: 3,566 out of 840 (maximum agreement: 752/840 for Llama 3 70B model)
- Inter-annotator agreement score: 0.7708 (strong level of agreement)
Evaluation of Pipeline on Previous Study Dataset
- Number of agreements between human annotators and voting classifier: 417 out of 600 comparisons
- Distribution of agreements per reproducibility variable:
- Model architecture: 89 agreements
- Open source framework: 53 agreements
Mapping of CQs to Reproducibility Variables in Previous Work
- Table 3 shows the mapping of CQs from this pipeline to the reproducibility variables in the previous work and number of agreements between human annotators from [^5] and voting classifier for each reproducibility variable.
Results after Filtering out Publications without DL in Study
- Number of publications after filtering: 257
- CQs mentioned before and after filtering:
- CQ 25 (purpose of the deep learning model): 345 mentions → 247 mentions
- CQ 27 (process to deploy the trained deep learning model): 6 mentions → 6 mentions
- Total number of queries answered: 3,524 out of 12,992 total queries
CQ Frequency in Remaining Publications after Filtering
| CQ Nr. | CQ | Number of publications that provide CQ info | Number of publications that provide CQ info after filtering the publications that do not contain DL in the study |
|---|---|---|---|
| 1 | What methods are utilized for collecting raw data in the deep learning pipeline (e.g., surveys, sensors, public datasets)? | 215/464 → 109/257 | |
| ... | ... | ... | ... |
| 27 | What is the process to deploy the trained deep learning model? | 6 → 6 |
Automated Approach for Retrieving DL Information from Scientific Articles
Challenges with Manual Approaches:
- Labour-intensive and time-consuming
- Variability in annotations based on the annotator's perspective
- Affects reproducibility of manually annotated data
Proposed Automated Approach:
- Employs five different Language Models (LLMs) to improve:
- Accuracy
- Diversity of information extraction
- Three critical components:
- Identifying relevant research publications
- Extracting relevant information from publications
- Converting extracted textual responses into categorical responses
Identifying Relevant Publications:
- Extracts publications based on selected keywords
- Derived from AI-related abstracts at Biodiversity Information Standards (TDWG) conference
- Total of 25 keywords
- Even if a publication mentions only one keyword, it will still be included in the extraction process
- After filtering, the number of publications decreased by 44.6% to 257
Evaluation:
- Achieved an accuracy of 93% when tested with 100 publications from previous work
- After filtering, positive response rate improved from 27.12% to 35.77%
- Still 64.23% of the queries did not yield available information in the publications
Extracting Relevant Information:
- Employed an RAG-assisted LLM pipeline to extract relevant information for all CQs and publications
- Generated a total of 12,992 textual responses across different LLMs
- High cosine similarity between Llama 3.1 70B - Llama 3 70B model pair
- Lower cosine similarity for Gemma 2 9B - Mixtral 8x22B Instruct v0.1 model pair
Converting Textual Responses to Categorical Responses:
- Facilitates evaluation process and creation of an ensemble voting classifier
- Two human annotators provided assessments for effective conversion
- Highest IAA scores observed for Llama 3 70B, Llama 3.1 70B, and Gemma 2 9B models
- Lowest IAA scores for Mixtral 8x22B Instruct v0.1 and Mixtral 8x7B models
Categorical Response Agreement:
- Moderate to strong agreement between various LLMs before filtering
- Improved after the publication filtering process
Comparison with Manual Annotations:
- Lowest agreement for datasets, open-source frameworks, and hyperparameters
- Differences in how dataset availability was considered
Voting Classifier:
- Enriches decision-making process and improves classifier's overall reliability
Evaluating Accessibility of Methodological Information in DL Studies
Background:
- Concern about lack of accessible methodological information in DL studies
- Systematic evaluation to address this issue
Approach:
- Generate machine-accessible descriptions for corpus of publications
- Enable authors/reviewers to verify methodological clarity
Applicability:
- Applies across scientific domains where detailed reporting is essential
- Previously demonstrated in biodiversity studies
Methodology:
- Automatic information retrieval through RAG-assisted LLM pipeline
- Llama-3 70B, Llama-3.1 70B, Mixtral-8x22B-Instruct-v0.1, Mixtral 8x7B, Gemma-2 9B used for ensemble result
- Comparison of outputs with human responses from previous work [^5]
Findings:
- Different LLMs generate varying outputs for the same query
- Precise indexing and filtering enhances results
- Incorporating multiple modalities improves retrieval process
Future Research:
- Develop a hybrid system combining human expertise with LLM capabilities
- LLMs evaluate results using metric to ensure accuracy
- Humans manually assess cases with low metric scores
- Include different modalities (code, figures) in pipeline for more accurate information retrieval.
This research is supported by iDiv and funded by DFG (FZT 118 & TRR 386). Computing time was granted through NHR@Göttingen's Resource Allocation Board for project nhr_th_starter_22233.
"The data and code for this study can be found on GitHub at https://github.com/fusion-jena/information-retrieval-using-multiple-LLM-and-RAG."