-
Notifications
You must be signed in to change notification settings - Fork 4
New issue
Have a question about this project? Sign up for a free GitHub account to open an issue and contact its maintainers and the community.
By clicking “Sign up for GitHub”, you agree to our terms of service and privacy statement. We’ll occasionally send you account related emails.
Already on GitHub? Sign in to your account
why using a 4d array in cp? #5
Comments
|
Hi @ccshao, Thanks for using scTenifoldNet. In the beginning, we tried to decompose the tensor with the two sets of networks at once, and established the structure of the code as a 4 order tensor. Then, our collaborators in statistics suggested that decomposing the tensor indepently for each set of networks was a most appropriate procedure. We keep the former structure and let the other dimension empty. I have modified the function to compute the 3d order tensor for you and I will check it out to see how it does affect the results of scTenifoldNet. Please let me know if there is something else I can help you with, Best, Daniel |
|
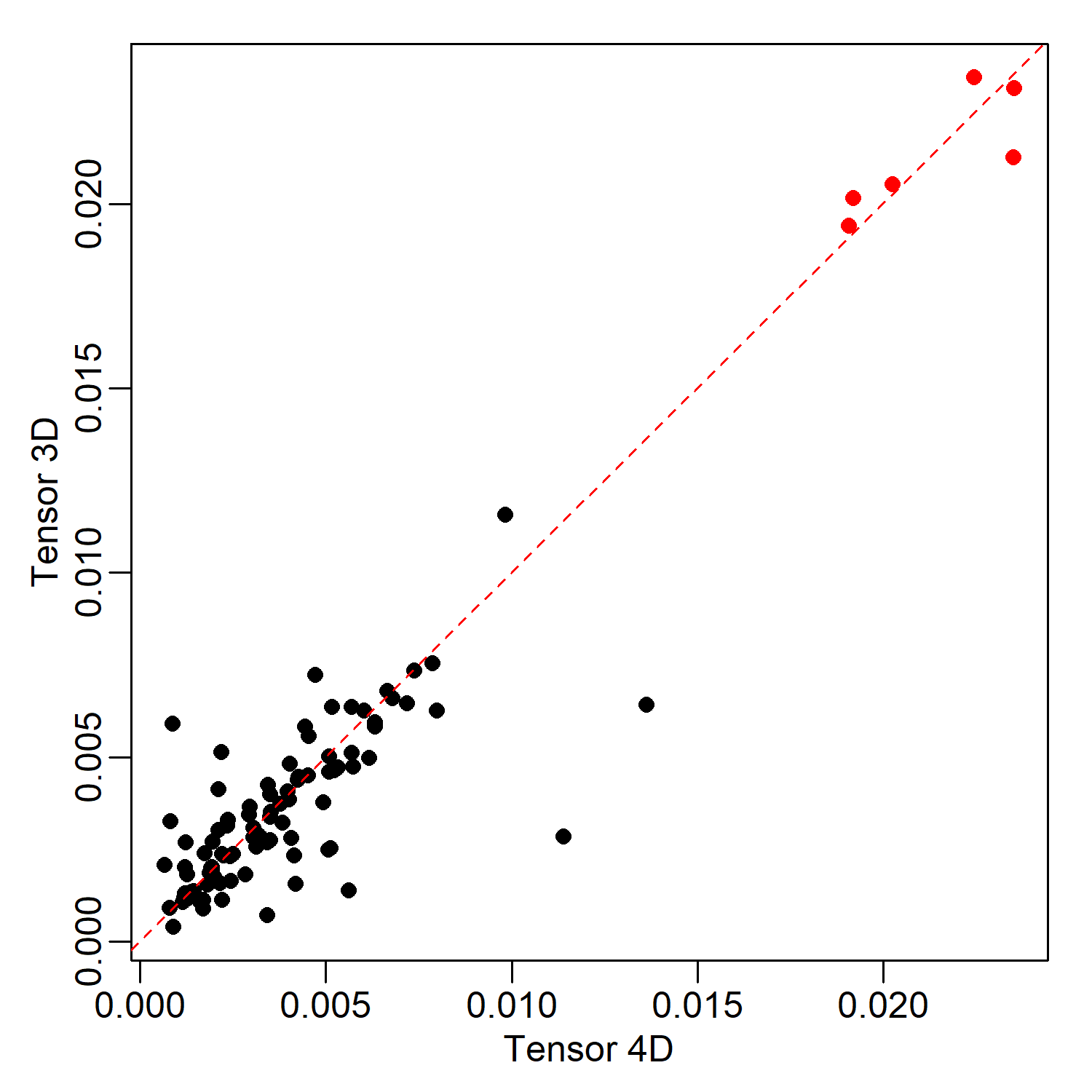
Hi @ccshao Thanks for pointing this out. I have tested the effect on the results of scTenifoldNet. I found that the identified genes as significant are the same, and the Pearson correlation coefficient between the distances is 0.941. Please see the code below: |
|
@dosorio Thanks very much for the quick updates, indeed it seems the choice of 4d/3d array makes little difference in comparing two networks. However, when I compared two networks from a single X datasets from the 4d/3d approaches, cp gives quite different structures indicated with low correlation value. In this scenario, which network is preferred? Note I used the cran version 1.0.0, but the tensorDecomposition2 is nearly identical to your codes in The codes used to produce a single network with either 3d or 4d. #- CP with 3d array |
|
Hi @ccshao, We have tested and benchmarked the 4d configuration against 11 other packages using Beeline and it ranks in first tied with PIDC for the network construction. Please see the attached figure: |
|
@dosorio would you mind to share the benchmark codes used in Beeline for the 12 methods? It would be interesting to see how the cp 3d array works. |
|
Hey @ccshao, Sure. All the required code is available at: https://github.com/cailab-tamu/scTenifoldNet/tree/master/inst/benchmarking/Beeline Best, Daniel |
|
Thanks! I will try to play with different settings of scTenifoldNet. |
|
Hi @ccshao, after the first release of scTenifoldNet, we received a notification from CRAN that the rTensor package was no longer maintained, and they planned to shut it down. We added the function as native in our package and added the citation to the original package. Nothing else changed. |


Dear scTenifoldNet dev,
Thanks very much for the interesting method. That is the first time I learn something about tensor low approximation,
ane I would like to try it on my work.
As I am new to the idea of tensor, I was confused on the 4d array used in the rTensor::cp steps. As the N * N * T (N is the number of genes, T is the sampling times) is 3d array, why a 4d array is used in the implementations (example codes taken from CRAN 1.0.0)?
Her are codes used In the script of tensorDecomposition.R,
Actually I have tried
tensorDecompositionwith 3d array, i.e., simly remove the 3rd dim; and replace the "1" in the 3rd to other values. In the former senario I got different results compared to your codes, and the latter case there was an error:Thanks in advance.
The text was updated successfully, but these errors were encountered: