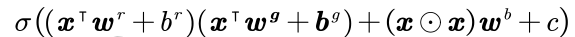This is the repository of our submission for IEEE Transactions on Neural Networks and Learning Systems "Heterogeneous Autoencoder Empowered by Quadratic Neurons". In this work,
- We develop a novel autoencoder that integrates essentially different types of neurons in one model;
- We present a constructive theorem to show that for certain tasks, a heterogeneous network can have more efficient and powerful approximation;
- Experiments on anomaly detection suggest that the proposed heterogeneous autoencoder delivers competitive performance compare to baseline.
All experiments are conducted with Windows 10 on an AMD R5 3600 CPU at 3.60 GHz and one NVIDIA RTX 2070S 8GB GPU. We implement our model on Python 3.8 with the PyTorch package, an open-source deep learning framework.
A quadratic neuron was proposed by [1], It computes two inner products and one power term of the input vector and integrates them for a nonlinear activation function. The output function of a quadratic neuron is expressed as
where
We propose three heterogeneous autoencoders integrating conventional and quadratic neurons in one model, referred to as HAE-X, HAE-Y, and HAE-I, respectively.

This repository is organized by three folders, which correspond to the three main experiments in our paper. Three programs (quadratic_autoencoder, ensemble_autoencoder, and heterogeneous_autoencoder) can be run independently.
We use PyCharm 2021.2 to be a coding IDE, if you use the same, you can run this program directly. Other IDE we have not yet tested, maybe you need to change some settings.
- Python == 3.8
- PyTorch == 1.10.1
- CUDA == 11.3 if use GPU
- pyod == 0.9.6
- anaconda == 2021.05
HAE_Empowered_by_Quadratic_Neurons
│ benchmark_method.py # Implementation of OCSVM, SUDO, DeepSVDD
│ train_ae.py # Implementation of AE\QAE\HAEs
│ train_DAGMM.py # Implementation of DAGMM
│ train_RCA.py # Implementation of RCA
└─ data # Anomaly detection datasets
└─ utils
│ data_process.py # data_process functions for RCA and DAGMM
│ QuadraticOperation.py # Quadratic neuron function
│ train_function.py # Train function for quadratic network
└─ Model
│ DAGMM.py
│ HAutoEncoder.py # Contain AE\QAE\HAEs
│ RCA.py
We use the ODDs dataset[2]. More details can be found in Official Page of ODDs Dataset.
Run train_ae.py to train an autoencoder. We provide three heterogeneous autoencoders, a quadratic and a conventional autoencoder.
Run /benchmark_method.py to train OCSVM, SUDO, DeepSVDD. We follow the implementation by pyod package [3].
Run /train_DAGMM.py to train DAGMM[4]. We follow the implementation by RCA.
Run /train_RCA.py to train RCA[5]. We follow the implementation by RCA.
All results will be saved to the 'results' folder.
The autoencoders we used in train_ae.py have been fine-tuning. The table below are the hyperparameters we recommend for traing. Note that RCA is also an autoencoder model, but authors use the same parameters on all datasets, so that we don't tune this model.
| datasets | model | batch size | learning rate | alpha |
|---|---|---|---|---|
| arrhythmia | AE | 128 | 0.00030 | - |
| QAE | 64 | 0.00008 | 0.15 | |
| HAE-X | 128 | 0.00008 | 0.05 | |
| HAE-Y | 128 | 0.00008 | 0.05 | |
| HAE-I | 128 | 0.00008 | 0.00 | |
| glass | AE | 128 | 0.00008 | - |
| QAE | 128 | 0.00100 | 0.35 | |
| HAE-X | 128 | 0.00030 | 0.00 | |
| HAE-Y | 128 | 0.00100 | 0.30 | |
| HAE-I | 128 | 0.00030 | 0.00 | |
| musk | AE | 32 | 0.00100 | - |
| QAE | 32 | 0.00100 | 0.00 | |
| HAE-X | 32 | 0.00100 | 0.00 | |
| HAE-Y | 32 | 0.00100 | 0.00 | |
| HAE-I | 32 | 0.00100 | 0.00 | |
| optdigits | AE | 64 | 0.00080 | - |
| QAE | 32 | 0.00080 | 0.35 | |
| HAE-X | 64 | 0.00008 | 0.30 | |
| HAE-Y | 128 | 0.00008 | 0.30 | |
| HAE-I | 128 | 0.00080 | 0.35 | |
| pendigits | AE | 128 | 0.00008 | - |
| QAE | 64 | 0.00100 | 0.15 | |
| HAE-X | 64 | 0.00008 | 0.20 | |
| HAE-Y | 64 | 0.00100 | 0.05 | |
| HAE-I | 128 | 0.00008 | 0.30 | |
| pima | AE | 32 | 0.00030 | - |
| QAE | 32 | 0.00080 | 0.15 | |
| HAE-X | 32 | 0.00100 | 0.05 | |
| HAE-Y | 32 | 0.00080 | 0.05 | |
| HAE-I | 64 | 0.00080 | 0.35 | |
| vertebral | AE | 128 | 0.00008 | - |
| QAE | 128 | 0.00008 | 0.25 | |
| HAE-X | 128 | 0.00008 | 0.25 | |
| HAE-Y | 128 | 0.00008 | 0.20 | |
| HAE-I | 128 | 0.00008 | 0.05 | |
| wbc | AE | 32 | 0.00008 | - |
| QAE | 32 | 0.00050 | 0.25 | |
| HAE-X | 32 | 0.00100 | 0.00 | |
| HAE-Y | 32 | 0.00100 | 0.00 | |
| HAE-I | 32 | 0.00100 | 0.20 |
Here we give the main results of our paper. We use AUC as a performance metric. All resutls are run 10 times to calculate the average. The proposed method outperforms other compared baseline methods.
if you have any questions about our work, please contact the following email address:
Enjoy your coding!
[1] Fenglei Fan, Wenxiang Cong, and Ge Wang. A new type of neurons for machine learning. International journal for numericalmethods in biomedical engineering, 34(2):e2920, 2018.
[2] Shebuti Rayana. Odds library [http://odds.cs.stonybrook.edu]. stony brook, ny: Stony brook university.Department of ComputerScience, 2016.
[3] Zhao, Y., Nasrullah, Z. and Li, Z., 2019. PyOD: A Python Toolbox for Scalable Outlier Detection. Journal of machine learning research (JMLR), 20(96), pp.1-7.
[4] B. Zong, Q. Song, M. R. Min, W. Cheng, C. Lumezanu, D. Cho, and H. Chen, “Deep autoencoding gaussian mixture model for unsupervised anomaly detection,” in International conference on learning representations, 2018.
[5] B. Liu, D. Wang, K. Lin, P.-N. Tan, and J. Zhou, “RCA: A Deep Collaborative Autoencoder Approach for Anomaly Detection,” pp. 1505–1511, 2021, doi: 10.24963/ijcai.2021/208.