Developed by the AIMS Lab at the Robarts Research Institute
2023–2025
⚠️ This package is under active development. While stable and reproducible, users are encouraged to report any bugs or unexpected behavior to the development team.
- 🧠 What is AutoAFIDs?
- ⚙️ Workflow Overview
- 🔍 Known Issues
- 📖 Full Documentation
- 💬 Questions, Issues, and Feedback
- 📚 Relevant Papers
AutoAFIDs is a BIDS App for automatic detection of anatomical fiducials (AFIDs) on MRI scans. We make use of these AFIDs in various neuroimaging applications such as image registration, quality control, and neurosurgical targeting.
AutoAFIDs leverages:
- A 3D U-Net architecture for landmark localization
- The Snakemake and SnakeBIDS workflow frameworks for reproducible processing
- The AFIDs protocol developed by the AFIDs community
The software is modality-aware, but best supports T1-weighted MRI scans. It also includes QC visualizations for quality assurance.
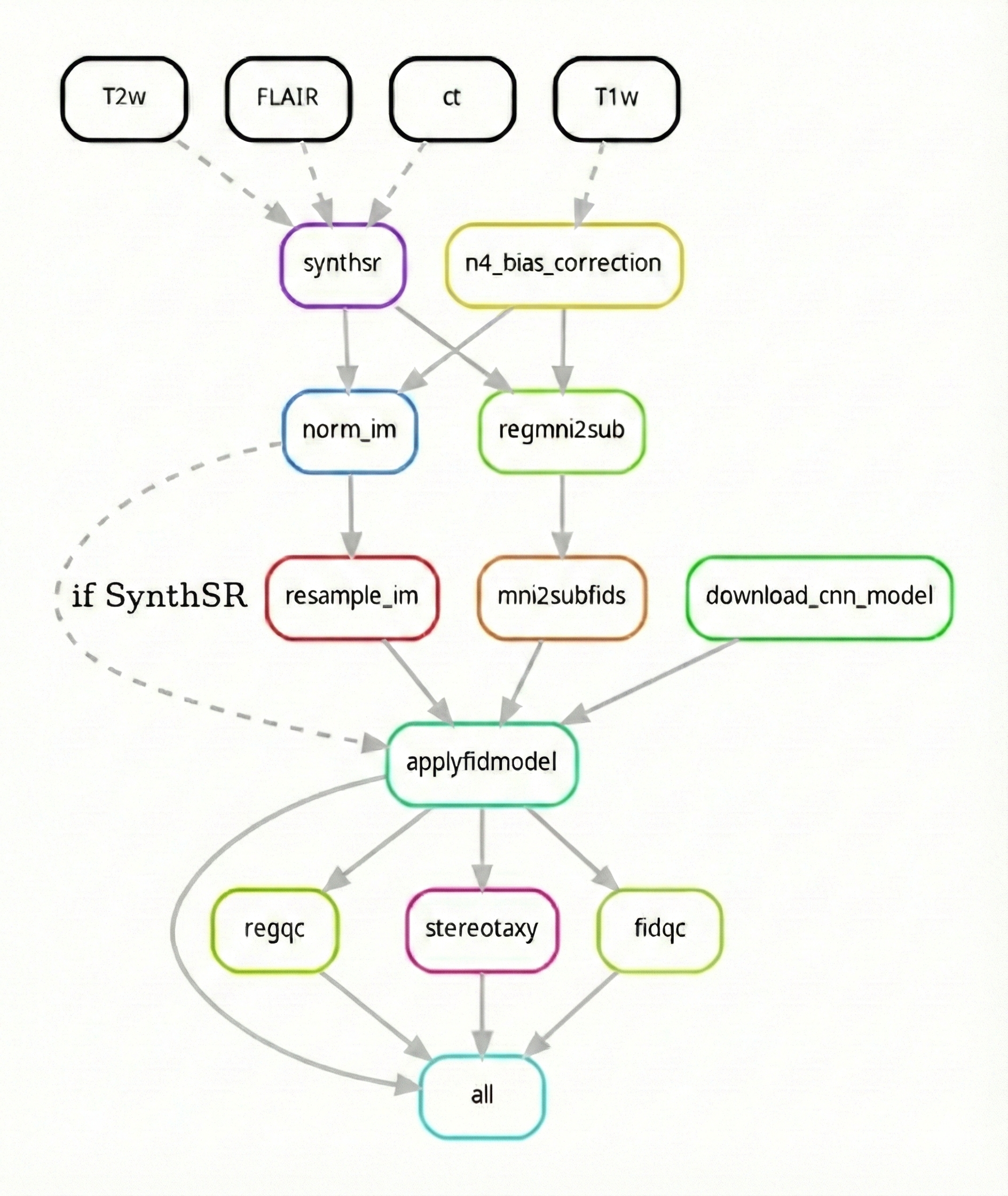
Below is a high-level summary of the AutoAFIDs processing pipeline:
- Preprocess BIDS input files based on image modality (T1w, T2w)
- Load trained fiducial models (32 AFID points)
- Run patch-based U-Net inference per AFID
- Generate predictions
gen_fcsvrule is currently sequential and may benefit from AFID-level parallelization- T1w-like scan synthesis is available (via SynthSR Iglesias et al., 2023) but requires millimetric validation and may not work for all operating systems
- AutoAFIDs does not currently filter for the
descentity in BIDS input files, since it outputs files with a fixed, customdescvalue. Therefore, BIDS input filenames must not include adescfield, or the pipeline may fail to resolve inputs correctly.
Includes installation instructions, usage examples, and advanced configuration.
- Open an issue on the GitHub issues page
- Start a conversation on the Discussions board
- Contact: ataha24@uwo.ca or dbansal7@uwo.ca
We welcome feedback, contributions, and collaboration!
AutoAFIDs builds upon a series of foundational works that introduce, validate, and apply the anatomical fiducials (AFIDs) framework for neuroimaging quality control, registration evaluation, and surgical planning.
- Lau et al., 2019
A framework for evaluating correspondence between brain images using anatomical fiducials.
Human Brain Mapping, 40(14), 4163–4179.
DOI: 10.1002/hbm.24695
-
Abbass et al., 2022
Application of the anatomical fiducials framework to a clinical dataset of patients with Parkinson’s disease.
Brain Structure & Function, 227(1), 393–405.
DOI: 10.1007/s00429-021-02408-3 -
Taha et al., 2022
An indirect deep brain stimulation targeting tool using salient anatomical fiducials.
Neuromodulation: Journal of the International Neuromodulation Society, 25(8), S6–S7.
- Taha et al., 2023
Magnetic resonance imaging datasets with anatomical fiducials for quality control and registration.
Scientific Data, 10(1), 449.
DOI: 10.1038/s41597-023-02330-9
- Abbass et al., 2025
The impact of localization and registration accuracy on estimates of deep brain stimulation electrode position in stereotactic space.
Imaging Neuroscience.
DOI: 10.1162/imag_a_00579
📌 If you use AutoAFIDs or AFIDs data in your research, please cite the relevant papers above.